Due to the frequent exposure to patients with COVID-19, health care workers (HCWs) are at high risk for SARS-CoV-2 infection. Presymptomatic or asymptomatic transmission account for around half of the cases. Therefore, increasing capacity and early diagnostic testing would help to reduce the transmission of the virus5. Screening approaches are mostly focused on symptomatic HCWs16 and there are few studies detecting SARS-CoV-2 in pre-symptomatic or asymptomatic HCW11,12.
The nasopharyngeal swab (NPS) is the most common respiratory sample utilized for SARS-CoV-2 diagnosis. How-ever, NPS is associated with subjects’ discomfort and further exposure to viral aerosols during sample collection8. Saliva testing is a non-invasive test to detect SARS-CoV-2 that avoids further HCW exposure. Saliva testing proved to be as sensitive as NPS in detecting SARS-CoV-2 in symptomatic patients6,7. However, the role of saliva testing among asymptomatic HCWs is still to be determined.
The objective of this study was to determine the performance of the saliva test to screen for SARS-CoV-2 in HCWs.
A descriptive prospective cohort study in HCWs was conducted at CEMIC University Hospital, Buenos Aires, Argentina. Voluntary SARS-CoV-2 testing in saliva was per-formed in both hospital sites: CEMIC Saavedra and CEMIC Pombo. HCWs were enrolled at 12 different hospital set-tings. These settings were classified as ''high’’, ''middle’’ or ''low’’ in terms of exposure risk to SARS-CoV-2. High exposure risk settings included: emergency room, inten-sive care unit (ICU), infectious diseases, and personnel assisting COVID-19 positive inpatients or specimens (tech-nicians, nurses, physicians, physical therapists, laboratory). Middle exposure risk settings included: personnel assisting COVID-19 negative inpatients (technicians, nurses, physicians, physical therapists). Low exposure risk areas included those without contact with patients or clinical specimens (pharmacy, administration).
An electronic form with personal data and information related to COVID-19 disease or exposure was collected at the time of sampling. Clinical follow-up for any signs or symptoms compatible with COVID-19 was obtained from par-ticipants for two weeks from the initial collection of saliva. A nasopharyngeal swab and a blood sample for serology were obtained from participants with positive RT-PCR results for SARS-CoV-2.
The study was approved by the CEMIC Ethics Commit-tee (Protocol: 1298/20) and electronic informed consent was obtained from all participants.
At study entry, a self-collected saliva sample was obtained for SARS-CoV-2 diagnosis. Participants were instructed on how to collect the sample in a sterile plastic container without viral transport medium. Samples were conserved at 4 °C until processed in a biosafety cabinet within 12 h of arrival. Viscous saliva samples were mechan-ically disrupted by adding 500 ^l viral transport medium [Minimun Essential Medium (Gibco); L-Glutamin 200 mM; HEPES 1 N; bovine serum albumin 5% (Sigma) sodium bicar-bonate 7.5%; penicillin, streptomycin and amphotericin (pH = 7.2)]. Participants in whom SARS-CoV-2 was detected by PCR in the initial sample of saliva were requested to pro-vide additional saliva samples and NPS. NPS were obtained and placed in a sterile tube containing 2 ml viral transport media.
Nucleic acid was extracted from 100 ^l saliva sample and eluted in 15 ^l using manual columns (Quick-RNATM Viral Kit. Zymo Research CORP.) following the manufacturer’s recommendation. One-step real-time multiplex RT-PCR laboratory-optimized was performed, targeting the SARS-CoV-2 E gene and the human RNAse P gene as quality control4,6. A positive result was considered when the cycle threshold (Ct) value for the SARS-CoV-2 E gene was lower than 40 and the human RNAse gene was positive. Analytical sensitivity was 1copy/^l. Discrepant PCR results were tested using a commercial RT-PCR kit that amplifies the SARS-CoV-2 E gene and the S gene and includes an internal amplification control (Real Star® SARS-CoV-2 RT-PCR Kit 1.0. Altona Diagnostics Argentina S.R.L.). Real time assays were performed in a CFX 96 Deep WellTM Real Time System (BioRad).
Serum samples were analyzed for the presence of SARS-CoV-2 specific IgM and IgG antibodies using ELISA kit COVIDAR IgM/IgG. The assay uses a trimer stabilized spike protein and the RBD (FIL-CONICET-Laboratorio Lemos, Argentina)9.
Ct values of saliva vs. those observed in NPS were compared with a paired Student’s t-test. A two-sided p-value of <0.05 was considered significant (GraphPad Prism 8.0.2, San Diego California, USA).
From September 9th to October 13th 2020, 100 asymp-tomatic/presymptomatic HCWs were enrolled in the study. Demographic characteristics were described as appropriate. Almost two thirds were female (67%) and the median age was 37 years old (IQR = 31-46). Around half of the partici-pants (53%) described having been in contact with COVID-19 patients at some point and 3.8% of these participants reported previous SARS-CoV-2 infection, but were negative in saliva or NPS sample at the time of this screening.
Of 100 HCWs, 6 asymptomatic subjects (6%) were pos-itive in saliva (2 physicians, 2 nurses, and 2 administrative personnel, respectively). All positive HCWs belonged to high exposure settings. Upon the initial results in saliva, all these HCWs were separated and licensed from work on the same day of sample collection.
NPS from positive patients were mostly obtained within 24-48 h from saliva testing. Five of six HCWs with positive salivawere positive in subsequent NPS (Table 1), no significant differences between Ct values (saliva vs. NPS) were observed (p = 0.3173). A patient with negative NSP showed a positive saliva test with a high Ct value. This result was confirmed in duplicate and also using a different RT-PCR assay (home-brew and commercial kit Altona Diagnostic). A follow-up saliva sample obtained 48 h later was also positive but with a higher Ct value. Among HCWs with positive saliva, 4/6 developed signs and symptoms compatible with COVID-19 within 2-4 days after the initial collection of saliva. The other 2 HCWs remained asymptomatic. Median Ct values in saliva were lower in pre-symptomatic (mean = 28.36; range: 15.26-36.80) versus asymptomatic (mean = 34.63; range: 33.71-35.53) HCW (p = 0.4033). Of 4 pre-symptomatic HCWs with positive saliva, 3 seroconverted. None of the two HCWs who remained asymptomatic seroconverted (Table 1). A follow-up saliva sample obtained from the asymptomatic HCW (ID #4) confirmed positivity although with a higher Ct value (Ct = 37). None of 94 HCWs who tested negative for SARS-CoV-2 in saliva developed symptoms.
Expanding screening protocols for detecting pre-symptomatic or asymptomatic carriers among HCWs is critical to reduce the spread of SARS-CoV-2 in the hospital setting. Some of the challenges to establish these proto-cols are related to logistical issues, turnaround times and further exposure to HCW during sampling. Self-collected saliva provides an option to facilitate sample collection, reduce discomfort and minimize HCW exposure. There are few reports utilizing saliva samples in the routine test-ing of asymptomatic or pre-symptomatic patients, including HCWs15. Generally, saliva tests were utilized for follow-up after a positive NPS but not as a primary screening method13. In this study, we have used saliva samples as a primary screening method to detect SARS-CoV-2 among HCWs. The incidence of SARS-CoV-2 in pre-symptomatic/asymptomatic
HCWs reaching 6% was higher than expected. Most of our positive cases corresponded to HCWs performing work activities in high or middle risk areas. Some authors reported negative results for saliva testing among HCWs1 while oth-ers found percentages ranging from 1.5 to 12.5% using NPS in those asymptomatic12,14,16. Our screening results in saliva are comparable with studies using NPS. A high correlation between saliva and NPS was previously demon-strated in symptomatic patients using a highly sensitive home brew RT-PCR6. In this study, saliva was a useful, valuable and easy tool for SARS-CoV-2 screening and for identifying pre-symptomatic and asymptomatic individuals. Importantly, rapid identification of positive HCWs permitted early licensing avoiding potential SARS-CoV-2 spreaders within the hospital setting. Furthermore, saliva positivity anticipated clinical disease up to 3 days before symptom onset. Long-term SARS-CoV-2 RNA shedding can occur in saliva. In our study, the only patient who gave a discrepant result between NPS and saliva and remained asymptomatic showed a follow-up saliva with a higher Ct value, probably representing prolonged shedding in this sample type17.
Of 6 positive HCWs, 2 remained asymptomatic. Studies evaluating nasopharyngeal swabs in HCWs demonstrated that 15% to 57% remained asymptomatic2,11. Seroconversion was demonstrated in most of our symptomatic patients. The patient who did not seroconvert even 90 days after symptom onset had a very mild disease. This finding is not unexpected since IgG titers have been associated with disease severity9. In addition, lack of seroconversion was previously described in 17% of HCWs infected with SARS-CoV-23.
Saliva is an easy to obtain sample that can be self-collected. It is especially convenient for screening in individuals without respiratory symptoms. Furthermore, special attention should be given to asymptomatic individ-uals who may spread the virus more easily due to higher human interaction than symptomatic patients10. Interest-ingly, previous studies with saliva testing have shown discrepant results. We believe the reason for such discrepancy is related to sample processing and the PCR techniques employed. We have systematically conducted and applied an optimized PCR (home brew). Our technique includes mechanical disruption of the saliva, no addition of any stabi-lizer or buffer and an increased concentration of magnesium (3.8mM) in the PCR mix as well as modifications in cycling conditions6. Therefore, the optimization of our PCR resulted in an increased analytical and clinical sensitivity. Further-more, this home brew assay incurred less costs than those related to commercial PCR kits (data not shown).
This study has several limitations. Our investigation was based on voluntary participation and this may have intro-duced some voluntary bias. In addition, other populations need to be tested to define the role of saliva to detect SARS-CoV-2 in symptomatic, pre-symptomatic or asymptomatic subjects. Finally, our sample size was not large. However, the number was deemed to be epidemiologically meaning-ful. Supporting this concept, we were able to detect 6% of HCWs infected with SARS-CoV-2 while they were pre-symptomatic/asymptomatic.
The screening protocol with the saliva sample test proposed in this work permitted rapid personnel isolation avoiding further transmission of the virus in the hospital setting.
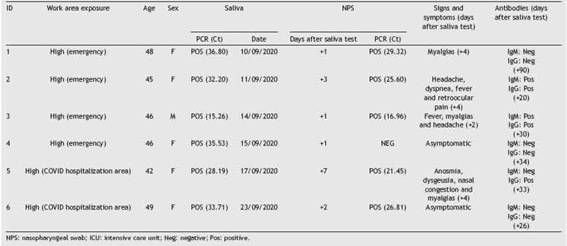
Table 1: Health care workers with a positive SARS-CoV-2 PCR in saliva. Clinical follow-up and NPS correlation.
FundingThis work was partially supported by a grant from Fondo para la Investigación Científica y Tecnológica (FONCYT) [IP-COVID19: 0938] awarded to Dr. Marcela Echavarría and by an internal fund from CEMIC and Fundación ''Norberto Quirno’’ [01/20].
Conflict of interestMS is a consultant to Basilea, a speaker for Pfizer and the principal investigator in Argentina for NIH grant UM1AI104681. The rest of the authors have no conflict of interest.
AcknowledgmentsWe are very grateful to all volunteers providing saliva sam-ples and permitting their clinical follow up. We specially thank Dr. Gamarnik and COVIDAR group for providing COVI-DAR ELISA kits and for their collaboration. We want to thank Maria Elvina Moussou, Fernando Lopes Mateo and Agostina Nicolelli for their help in sample storage and transportation. We also want to thank technician Victoria Cenal for helping in serum samples separation.












 uBio
uBio 

