INTRODUCTION
Currently, the Chaco vegetation is undergoing widespread and increasing desertification, resulting from the exploitation of natural resources, mainly from deforestation (Pasig, 2005). Past changes, aggravated by the loss of tree cover since 2000 and agricultural expansion, have made Chaco deforestation rates one of the highest in the world (Hansen et al., 2013; Vallejos et al., 2015; Baumann et al., 2017). This situation poses challenges to the Chaco region’s remnants, as fragmentation may affect the genetic diversity of populations (Hamrick, 2004).
In the past two decades, the significant changes of landscape and the evolutionary dynamics of natural populations under fragmentation have been studied in the tropics but mainly concerning rainforests (Jakovac et al., 2015), particularly in studies of adaptation, eco-evolutionary dynamics, and fragments (Hill et al., 2011; Lôbo et al., 2011; Taubert et al., 2018). Thus, most of the Chaco forest and savanna biomes, and other biogeographic regions with similar phytophysiognomies, are not considered in such studies but are indispensable for understanding the effects of fragmentation on species conservation and speciation process in the region (Werneck 2011; Hughes et al., 2013).
The effects of habitat fragmentation are documented as causing inbreeding and reducing the gene flow in natural populations, accelerating the reduction and extinction of these populations by reproductive isolation (Young & Boyle, 2000). Moreover, populations are at risk of compromising the colonization process in other areas due to decreased genetic variability by inbreeding (Young & Boyle, 2000; Schneider et al., 2003). These characteristics are crucial for the development and survival of individuals after possible regeneration and may determine a species’ population structure (Medeiros et al., 2007).
When exposed to disturbance, natural forest populations tend to restructure as they are modified, restoring breeding processes such as pollination, seed dispersal, and seedling recruitment (Cullen Júnior et al., 2006), and genetic variability, mainly through gene flow, which is necessary for the maintenance of populations (Futuyma, 1992). However, even if the identity and population function are maintained (Walker et al., 2004), the original genetic structure’s recovery is not guaranteed, as genetic diversity may vary if populations are persistent or colonizing.
Aspidosperma quebracho-blanco Schltdl. (Apocynaceae) is characteristic of the Chaco environment in Argentina, Brazil, Bolivia, Paraguay, and Uruguay (Marcondes-Ferreira & Kinoshita, 1996; Galvani & De Moura, 2003; Jørgensen et al., 2014). This species has a history of large-scale logging due to the presence of tannin in its wood, used in the manufacture of chemical products and leather tanning. These features motivated the installation of industries in the Chaco region, threatening the natural populations of A. quebracho-blanco (Hueck, 1955; Bandeira, 1998). Habitat disturbance and the restricted distribution of species are characteristics that may contribute to the evolution of self-compatibility systems, which initially may lead to low genetic variability and, consequently, niche amplitude, a factor of significant influence on the occupation of new areas (Lowry & Lester, 2006). Such conditions may be occurring in A. quebracho-blanco populations due to the fragmentation of their natural habitats.
Genetic diversity assessments of species threatened by habitat fragmentation have shown promising results when used in conservation strategies and the maintenance of natural populations (Ferreira & Grattapaglia, 1998; Finger et al., 2014). Genetic analysis provides data for better understanding patterns of the structure and distribution of genetic variability, which are important indicators for understanding evolutionary processes in natural populations. Studies using genetic data may also suggest areas for conserving a significant portion of genetic diversity, indicating the need for the creation of units of conservation. (Montagna et al., 2012).
Advances in molecular biological studies, such as DNA molecular markers for endangered species, facilitate conservation strategies and the use of genetic resources more appropriately (Avise, 2010; Frankham, 2010). Thus, conservation strategies that use these molecular markers can quantify the potential genetic resources available in natural populations, allowing us to elucidate and predict future genetic events, as well as connection points between populations and their evolutionary events (Cruz et al., 2011; Rajwant et al., 2011).
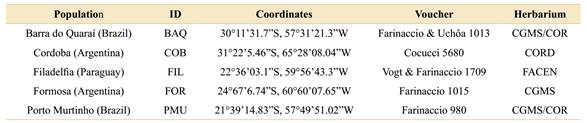
Table 1 Data of sampled populations of Aspidosperma quebracho-blanco. Geographic origins and voucher information from each population analysed in the present study.
Among the types of molecular markers used in population genetics analysis, ISSR (Inter Simple Sequence Repeats) has been increasing in popularity due to its effectiveness in detecting polymorphisms and genome abundance, low cost, and because it is a relatively simple technique (Gupta et al., 1994; Santana et al., 2011; Ng & Tan, 2015). Although ISSR markers have some limitations regarding low reproducibility and homology of co-migrating amplification products, these markers also use a single primer to obtain fragments and access germplasm fingerprinting of cultivated and native species, helping to elucidate evolutionary relationships among species (Manel et al., 2003; Faleiro, 2007; Amom & Nongdam, 2017). The specifics of each population obtained from molecular markers can be quantitatively related to geographical characteristics (Manel et al., 2003). Thus, this study aimed to present initial genetic structuring data of A. quebracho-blanco natural fragmented populations to suggest priority populations and guidelines, and for conservation and management efforts.
MATERIALS AND METHODS
Study area and sampling
Collection expeditions were carried out on three out of five known distribution areas of Aspidosperma quebracho-blanco (Argentina, Brazil, and Paraguay). All collected material was deposited in the herbaria of the Federal University of Mato Grosso do Sul (CGMS and COR), Cordoba Botanical Museum (CORD), and National University of Asunción (FACEN) (Table 1; acronyms following Thiers, 2020). The collected leaves were stored in silica gel for rapid dehydration (Chase & Hills, 1991). Overall, 25 individuals were collected, five samples from five populations: BAQ (Barra do Quaraí, Brazil), COB (Cordoba, Argentina), FIL (Filadelfia, Paraguay), FOR (Formosa, Argentina), and PMU (Porto Murtinho, Brazil) (Fig. 1).
The COB, FIL, FOR, and PMU populations were located in the Chaco area, an environment that has been disturbed in the past by logging and pasture clearing (Vallejos et al., 2015). In this biogeographic region, there is a more significant number of A. quebracho-blanco populations, unlike in Barra do Quaraí (Brazil), where there is only one population. The phytophysiognomy present in the Barra do Quaraí municipality is predominantly Steppe Savanna, with many fields of “espinilhos”. “Espinilho” is the popular name given to Vachellia (=Acacia) caven (Molina) Seigler & Ebinger, one of the most representative tree species in the region (Galvani & De Moura, 2003).
DNA extraction and loci selection
Genomic DNA was extracted from leaves according to the protocol of Doyle and Dickson (1987) with changes to the amount of isopropanol (750 µL) after collecting the aqueous phase. Extracted DNA was quantified on a 1% agarose gel stained with SyberGold ™ (Invitrogen®, Thermo Fisher Scientific, Waltham, Massachusetts) and compared to DNA of phage λ.
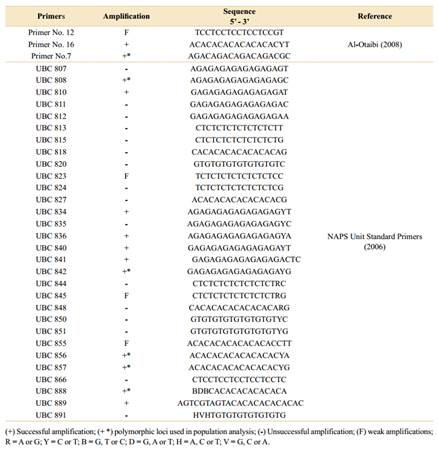
Thirty-three ISSR primers were tested, 30 of which were developed by the University of British Columbia Biotechnology Laboratory (UBCBL), named UBC primers (NAPS, 2019), and three others by Al-Otaibi (2008), named Primers Numbers 7, 12 and 16 (Table 2).
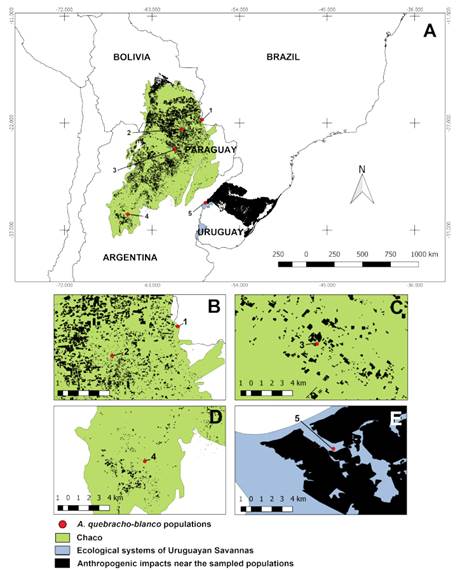
Fig. 1 Distribution map of the sampled populations in the Chaco vegetation area, Uruguayan Savannas’ ecological systems, and anthropization nearby. A, mapping of anthropization near the Steppe Savanna and deforestation in the Chaco, related to the sampled populations: PMU (1); FIL (2); FOR (3); COB (4) and BAQ (5). B-E, detail of impacts close to the sampled populations. Map made with QGIS® 3.0 software (QGIS, 2011) based on Veloso et al. (1991), Olson et al. (2001), and Hasenack et al. (2010). Color version at http://www.ojs.darwin.edu.ar/index.php/darwiniana/article/view/932/1213
Amplifications were performed by PCR in 25 µL reactions containing ≈30 ng DNA, 1X enzyme buffer, 2.0 mM MgCl2, 0.15 mM dNTP mix, 0.5 pmol ISSR primer, and 1 U of Taq DNA polymerase (Go Taq, Promega ©, Madison, Wisconsin, United States), amplified in a Veriti 96-well thermocycler (Applied Biosystems, Foster City, CA, USA) using the conditions previously described (Ribeiro et al., 2013; Vieira et al., 2014). Amplified products were visualized on SyberGold™ stained 1.5% agarose gel (Invitrogen ©, Thermo Fisher Scientific, Waltham, Massachusetts) and 1X TBE under a 120mA direct current. The molecular weight marker used was ladder 1 Kb (Invitrogen ©, Thermo Fisher Scientific, Waltham, Massachusetts), and the products were photographed with the incidence of blue light.
Genetic data analysis
To characterize the genetic diversity of A. quebracho-blanco populations, polymorphic ISSR loci with the largest number of fragments and reasonable resolution, and therefore more information, were selected. The polymorphic loci obtained were observed and photographed on agarose gels for a categorical analysis in ImageJ® software (Schneider et al., 2012). The collected data were used to construct a binary matrix based on the presence (1) and absence (0) of bands for all selected loci. The matrix was used to calculate the similarity matrix, considering the Jaccard index (Jaccard, 1908) as the arithmetic complement, with which genetic similarities were obtained within and between populations.
To establish a genotype grouping based on genetic similarity and construct a dendrogram, the unweighted pair group method (UPGMA) with the arithmetic mean was used (Sokal & Michener, 1958). The distance matrix and the dendrogram were obtained using the Past 3.0® program (Hammer et al., 2001). To analyse diversity within and among the studied populations, the Genes® program (Cruz, 2019) was used, estimating the following diversity parameters: Nei (He) genetic diversity (Nei, 1973), Shannon (H’) index (Shannon, 1948), polymorphic information content (PIC) ranging from 0 to 0.5 in dominant markers (Botstein et al., 1980) and heterozygosis, which was obtained from binary data (Cruz, 2019). The hypotheses of genetic data being differentiated because of distance isolation were also tested, and a correlation was established between geographic data and genetic distance matrices (FST/ (1-FST) by the Mantel test (Sokal & Rohlf, 1995) using GenePop On the web 4.2 (Raymond & Rousset, 1995).
Identification of fragmented habitats by georeferencing collection areas
The collected populations were represented cartographically using QGIS® 3.0 software, with the Chaco areas and ecological systems of Uruguayan Savanna limitations previously defined by Veloso et al. (1991) and Hasenack et al. (2010). The QGIS® 3.0 software was also used to identify landscape degradation that could pose potential risks to the sampled populations, using cartographic objects and collection locations represented with Chaco deforestation areas line-type shapefiles (Guyra Paraguay, 2019) and anthropization near the “espinilho” fields (“Projeto Monitoramento do Desmatamento dos Biomas Brasileiros” - Deforestation Monitoring Project for Brazilian Biomes).
RESULTS
Success of amplification and polymorphisms
The amplified products presented good electrophoretic profiles, totalling 450 bands in the 200 to 5000 bp regions. Of the 33 primers tested in A. quebracho-blanco, 16 were amplified (48%) (Table 2), with six selected loci presenting the best band patterns and polymorphisms under the conditions tested. In addition, 124 loci (two to six bands per population) were amplified, and of those, 37 polymorphisms (45.8%) were identified, with an average of six polymorphisms per marker. Among the loci selected for diversity and similarity analyses, an average of 20 sequences per primer were observed: 22 sequences (18.3%) in UBC 888, 21 (17.5%) in UBC 808 and UBC 856, 19 sequences (15.8%) in primer No.7 and UBC 842 and, 18 sequences (15%) in UBC 857.
Population similarities and geographic correlations
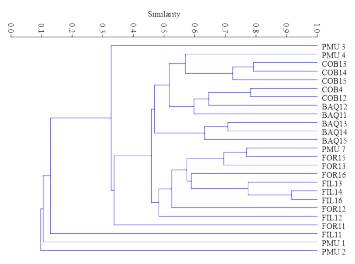
The coefficient values of the similarity matrix among populations ranged from 0.26 to 0.55, with a mean of 0.41; the highest similarity was between the FIL and FOR populations. The UPGMA-based dendrogram between the analysed population genotypes separated PMU from the other populations and formed two more groups with similarity values higher than 0.51. In branch number I, FOR and FIL populations were grouped, while in branch II a group with COB and BAQ was formed (Fig. 2).
Among the individuals sampled in different populations, the similarity matrix values ranged from 0.0 to 0.77, with values between individuals from the same population ranging from 0.0 to 0.92 (Table 3).
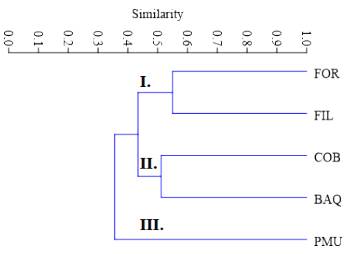
Fig. 2 UPGMA dendrogram of A. quebracho-blanco populations sampled, considering the Jaccard index as an arithmetic complement. Branch I: (FOR and FIL), Branch II: (COB and BAQ), Branch III (PMU).
Besides, according to the same criteria used for population analysis, the dendrogram of the 25 individuals analysed grouped representatives from different populations in close branches, with PMU being the population that presented the most distinct values of similarity among their individuals (Fig. 3). The Mantel test was not significant (r2 = 0.25, P = 0.106), showing no association between genetic and geographic distances, indicating the absence of isolation by distance between the sampled locations.
Genetic diversity
The mean genetic diversity observed in the populations ranged from 0.09 to 0.15 for the Shannon index and from 0.19 to 0.31 for Nei diversity (Fig. 4). Allele frequency and heterozygosis showed relatively close values in the FOR, FIL, COB, and BAQ populations (Table 4), coinciding with the PIC patterns obtained with the accessions of the six markers tested (Table 5).
Population distribution and environmental impacts
The population distribution was concentrated, that is, closer to each other, under the dry Chaco area (north of the sampled phytophysiognomies), by passing the wet Chaco formations until reaching the disjunction of the savanna vegetation classified as “fields with espinilho in the Steppe Savanna” (Fig. 1A). Higher environmental impacts were observed in the northern Chaco region and the Steppe Savanna than in other regions, where the populations of PMU, FIL, FOR, and BAQ are located (Fig. 1A-E). Georeferencing analyses at a smaller scale showed that the PMU population is in the Chaco border region also affected by deforestation; we also observed deforested regions very close to FIL and anthropized areas around Espinilho State Park (Brazil) in the BAQ population. However, the COB population was observed in conservation units within Chancaní Provincial Park’s limits (Argentina).
DISCUSSION
In agreement with our study, other works that used molecular markers in species of Aspidosperma Mart. & Zucc. showed the occurrence of values below 40% both in the number of amplified sequences and in the polymorphisms found by primers (Damasceno et al., 2011; Ferreira-Ramos et al., 2011; Zimback et al., 2011). In addition to this problem, which involves increased spending and time to obtain the results, DNA extraction from Aspidosperma leaves may be compromised due to the presence of a wide variety of alkaloids (Schumutz, 1960; Molinari & Crochemore, 2001; Sahu et al., 2012), and polysaccharides (Freitas, 2008), which are responsible for possibly inhibiting Taq DNA polymerase in the PCR (Demeke & Adams, 1992; Fang et al., 1992). Besides, added to the possibility of the occurrence of these substances in the DNA extraction, A. quebracho-blanco is a source species of tannins (Becker & Makkar, 1999), compounds that precipitate with the DNA, thus degrading DNA quality and reducing the yield (Katterman & Shattuck, 1983; Sarwat et al., 2006). Considering the possible implications of the DNA extraction and amplification procedures, we concluded that the products obtained were as expected and answered the proposed questions.
The spatial genetic structure observed in the A. quebracho-blanco natural populations represents the specific patterns of the spatial distribution of alleles and genotypes of individuals within populations (Vekemans & Hardy, 2004). Genetic analysis showed that the distribution of A. quebracho-blanco genotypes did not occur randomly, since we verified the presence of individuals from nearby sites in equal branches on UPGMA dendrogram (Fig. 3), there is a need for a new larger sample of individuals so that distance isolation can definitely be suggested (Wright, 1943). It is possible that this feature can be associated with seed dispersal and seedling recruitment, which could be favoured in the PMU, FIL, and FOR populations since the arid and semiarid Chaco environment is characterized by rainfall scarcity (Riveros, 2005), which favours anemochory (Howe & Smallwood, 1982), increasing the genetic similarity between these fragments due to gene flow via seeds (Hamrick et al., 1993). It is worth mentioning that this is a first approximation of the genetic structure of the sampled populations, with the need for an in-depth understanding of the eco-evolutionary processes of A. quebracho- blanco, whose limitations and inferences were not addressed by the ISSR techniques used here.
Geitonogamy is effective in A. quebracho- blanco, indicating that the species is self- compatible (Lin & Bernardello, 1999), a characteristic already reported in Apocynaceae species (Vieira & Grabalos, 2003; Löhne et al., 2004; Darrault & Schlindwein, 2005). The likelihood of foreign pollen arriving is only higher than the probability of self-pollination due to protandry and the position of the flowers (pending and sometimes in the horizontal position), characteristics that prevent self-fertilization (Lin & Bernardello, 1999). However, variations in the reproductive system, like self-compatibility, may reduce heterozygosity (Pollak, 1987; Schoen & Brown 1991) and possibly genetic diversity in natural populations (Hamrick & Godt, 1996; Glémin et al., 2006). Such variation may also partly justify the values obtained for the frequency of alleles, low heterozygosis, and polymorphic information (PIC) in this study.
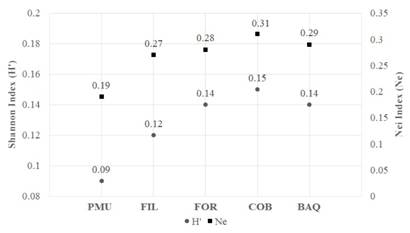
Fig. 4 Shannon index (H’) and Nei (Ne) genetic diversity for the five native populations of A. quebracho-blanco. Porto Murtinho, Brazil (PMU); Filadelfia, Paraguay (FIL); Formosa, Argentina (FOR); Cordoba, Argentina (COB); and Barra do Quaraí, Brazil (BAQ).
The establishment of autogamous populations is related to reductions in natural habitats, making cross-pollination difficult and guarantees significant survival of self-compatible mutants (Kaye, 1999). Similar effects have already been observed in plant reproductive systems from the Chaco Serrano (Aizen & Feinsinger, 1994), as is the case of A. quebracho-blanco, whose reproductive system is characterized by limited moth nocturnal pollination, and moths’ disorientation (Lin & Bernardello, 1999).
The northern and northwestern portions of the Chaco are characterized by sudden changes in land cover (Caballero et al., 2014), where the lowest values of genetic diversity were observed in the population. Notably, the areas around the BAQ population have experienced periods with variations in their hydromorphic soils due to degradation of surface horizons and natural regeneration of portions with signs of anthropogenic degradation (Da Cunha et al., 2001; Caldas et al., 2013). Thus, the sampled populations are surrounded by areas under anthropogenic impacts that threaten their current genetic diversity.

Table 3 Coefficient of the similarity matrix among the sampled individuals, considering the Jaccard index (Jaccard, 1908) as an arithmetic complement.
Likewise, the results presented here, habitat loss by anthropization, may also threaten other Apocynaceae tree species’ genetic diversity. In a study with Hancornia speciosa Gomes in fragmented habitats, for example, the Shannon index (H’) the Nei (He) index values were 0.26 and 0.18, respectively (Costa et al., 2015), which should be considered when establishing new conservation units of such populations with historical fragmentation and disturbance, which can make populations more vulnerable to the effects of genetic drift. Moreover, due to the greater likelihood of self-fertilization and interbreeding between closely related individuals, there is a possibility of inbreeding and border effects (Hartl & Clark, 1997; Kageyama et al., 1998).
Environmental conditions in arid locations are critical factors for seedling establishment (Watson et al., 1997; Dodd & Donovan, 1999). For regeneration of these regions, seedlings can be recruited from conservation units, as has been observed in seedlings from the Provincial Park Chancaní (Argentina) (Barchuk & Díaz, 1999), where the population of the highest genetic diversity was observed (COB). These data show the importance of comparing the genetic information with other data, and it is possible to infer issues involving management, recovery of degraded areas, and distance parameters for germplasm collection to obtain higher levels of genetic variability from the sampled populations (Cloutier et al., 2007).
Conservation actions are primarily necessary for the Porto Murtinho (PMU) population, as the region has experienced the most fragmentation among the known Chaco formations in recent decades (Zak et al., 2004; Grau et al., 2005). Furthermore, there are indications that this population’s genetic variability is similar to that in the other populations, showing similar genotypes. Along with the geographical distribution, the remaining Chaco populations show greater genetic diversity values in a north-south diversity gradient, i.e., the starting point that can be considered for genetic diversification of A. quebracho-blanco probably is PMU. Therefore, the Porto Murtinho population may present priority conditions for creating conservation areas, ensuring gene flow and genetic diversity among the other fragments (Vieira & Carvalho, 2008). Besides, it would be essential to construct a germplasm bank for the Cordoba (Argentina) population, which is a population with higher genetic diversity in a region of lower fragmentation than the other regions evaluated, in order to ensure the conservation of A. quebracho-blanco.

Table 4 Frequency values of alleles obtained and heterozygosis (H) for Aspidosperma quebracho-blanco populations sampled according to Genes® software (Cruz, 2019).
The estimates of genetic parameters have shown the need for the creation of protected areas for A. quebracho-blanco. Likewise, ecological aspects, such as the documentation of more than 200 kinds of insects associated with the availability of exudate extrafloral of this species (Lin & Bernardello, 1999), also suggest special attention to their ecological relationships is needed, as habitat degradation due to urbanization may play a role in altering arthropod diversity (Gibb & Dieter, 2002).
Our findings on the studied populations, considering the first genetic data and the geographical distribution of A. quebracho-blanco, indicate a possible threat to the genetic diversity of populations, an issue to be considered for the inclusion of A. quebracho-blanco on lists of threatened species, which are essential tools for planning conservation actions and programs (Mace et al., 2008). The results obtained here point out the importance of conservation units in maintaining the genetic diversity of natural populations, representing a solution for the conservation of A. quebracho- blanco genetic resources, since the damage caused by the anthropization of the Chaco and fields with “espinilho” may result in the loss of habitat and genetic diversity in the studied populations. Therefore, changes in the species conservation status and the creation of germplasm banks of populations from regions with minimal fragmentation effects are necessary, ensuring the conservation and reproduction of plant material as a safety mechanism for species.
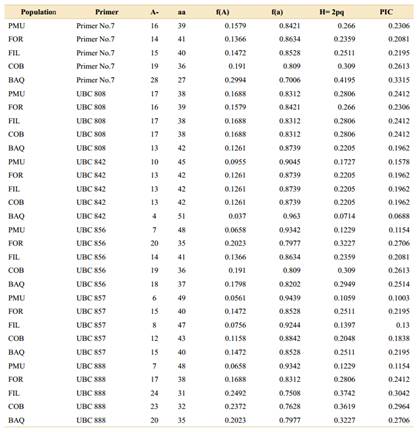
Table 5 Polymorphic information content (PIC) standards ranging from 0 to 0.5 for dominant markers (Botstein et al., 1980) and heterozygosis from binary data obtained with the information of the six markers in Aspidosperma quebracho-blanco populations, tested in the Genes® software (Cruz, 2019).












 uBio
uBio