Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
Darwiniana, nueva serie
Print version ISSN 0011-6793On-line version ISSN 1850-1699
Darwiniana, nueva serie vol.5 no.1 San Isidro July 2017
SISTEMÁTICA Y TAXONOMÍA DE ALGAS Y HONGOS
Trichoderma species associated with acromyrmex ant nests from Argentina and first report of trichoderma lentiforme for the country
Natalia G. Armando, Jorge A. Marfetán & Patricia J. Folgarait.
Laboratorio de Hormigas, Departamento de Ciencia y Tecnología, Universidad Nacional de Quilmes, Roque Sáenz Peña 352, B1876BXD Bernal, Buenos Aires, Argentina; ng.armando14@gmail.com (author for correspondence), pfolgarait@unq.edu.ar (author for reprints).
Abstract
The aim of this work is the morphological and molecular identification of five species of Trichoderma associated with leaf-cutting ant nests of the genera Acromyrmex present in Argentina. The species identified were T. lentiforme, T. inhamatum, T. virens, T. koningiopsis and T. aff. neotropicale resulting in the first record of T. lentiforme and T. inhamatum associated with leaf-cutting Acromyrmex ants, in particular with Acromyrmex lobicornis and Acromyrmex lundii for the first one and with A. lobicornis for the second one. Moreover, T. lentiforme represents the first record for Argentina. In this work we extend the measurements of the conidia and the conidiophore and contribute with additional of the distribution of the species of Trichoderma in this country. Photographs illustrating conidiophores, conidiogenous cells, conidia, and colony phenotype are provided for each species.
Keywords: Ascomycota; Leaf-cutting ants; Molecular identification; Morphological identification; Taxonomy; Tef1.
Especies de Trichoderma asociadas con nidos de hormigas del género Acromyrmex en Argentina y primer registro de Trichoderma lentiforme para el país
Resumen
Este trabajo tiene como objetivo la identificación morfológica y molecular de cinco especies de Trichoderma asociadas con los nidos de hormigas cortadoras de hojas del género Acromyrmex presentes en Argentina. Las especies identificadas fueron: T. lentiforme, T. inhamatum, T. virens, T. koningiopsis y T. aff. neotropicale resultando éste el primer registro de T. lentiforme y T. inhamatum asociados a hormigas cortadoras de hojas Acromyrmex, en particular Acromyrmex lobicornis y Acromyrmex lundii para el primero y de A. lobicornis para el segundo. Además T. lentiforme representa el primer registro para la Argentina. En este trabajo se amplían medidas del conidióforo y conidios y se aportan datos adicionales sobre la distribución de las especies de Trichoderma en el país. Se ilustran con fotografías los conidióforos, células conidiógenas, conidios y el fenotipo de la colonia para cada especie.
Palabras clave: Ascomycota; Hormigas cortadoras de hojas; Identificación molecular; Identificación morfológica; Taxonomía; Tef1.
Original recibido el 17 de agosto de 2016,
aceptado el 14 de julio de 2017
Editor Asociado: Mario Saparrat
INTRODUCTION
Leaf-cutting ants (Hymenoptera: Formicidae: Attini) are considered to be the major herbivores in the Neotropics, distributed exclusively in the New World. These ants can be grouped into two genera, Atta and Acromyrmex (Holldobler & Wilson, 1990). Leaf-cutting ants cut fresh vegetable material and use it as substrate for a symbiotic fungus, which provides a nutritional source used to feed the queen and the larvae (Weber, 1972). Ants cultivate species from the genus Leucoagaricus, such as L. gongylophorus and L. weberi, as well as other undefined morphotypes from the same genus (Singer, 1986; Muchovej et al., 1991; Folgarait et al., 2011; Lugo et al., 2013).
Although the fungal growth used to be considered a monoculture over a vegetable substrate derived from a wide variety of plant species, recent research has shown that the situation is more complicated. The Attini fungal garden is continuously exposed to alien microorganisms (Möller, 1893; Fisher et al., 1996; Rodrigues et al., 2005, Montoya et al., 2016) giving place to a microbiota complex associated with the leaf-cutting cultivar (Scott et al., 2010). Within this microbiota several groups of microorganisms can be found, such as yeasts (Carreiro et al., 1997; Pagnocca et al., 2001; Little et al., 2006), bacteria (Currie et al., 1999a; Pinto-Tomás et al. 2009), and anamorphic fungi (Currie et al., 1999b; Rodrigues et al., 2005; Ribeiro et al., 2012), some of which are saprophytes, nitrogen fixators, entomopathogens or pathogens of Leucoagaricus spp. Fungi associated with the cultivar, or mycobiota, are actually a very complex community with a lot of species growing inside the ant nest interacting in several ways.
The genus Trichoderma (Ascomycota: Hypocreales) contains cosmopolitan and ubiquitous species associated with a wide variety of substrates. Species of this genus can be usually found in soil, rotting plant material, other fungi, and as endophytes in the sapwood of tropical trees (Chaverri & Samuels, 2003; Samuels, 2006; Jaklitsch, 2009). Additionally, the genus Trichoderma was also isolated growing in the fungal garden of Acromyrmex species (Rodrigues et al., 2008).
Recent studies using different molecular markers (Samuels & Ismaiel, 2009; Druzhinina et al., 2011) revised the taxonomy of the genus Trichoderma as well as of T. koningii and T. harzianum species complex, describing new species (Samuels et al., 2006; Chaverri et al., 2015). Several of these species were isolated from different sites across the world, such as Cameroon, Sri Lanka, USA, Italy, Germany, England, France, Austria, Greece, Croatia, Spain, Mexico, Ecuador, Brazil, Peru, Japan, Ireland and China. There is little knowledge about the Trichoderma species present in Argentina. The most relevant data about this genus was collected by Barrera (2012), responsible for the most thorough research in Argentina: 38 species were found growing on roots, fallen leaves, decaying wood substrate and unproductive soil, 17 of them were new records in Argentina. Similarly, there is scarce information of Trichoderma species within leaf-cutter ant nests (Rodrigues et al., 2008). A recent study, containing the largest sampling of Trichoderma related to Attini ant nests from Brazil and United States of America, found 20 species of this genus, three of which were new species (Montoya et al., 2016). Still, Trichoderma species associated with leaf-cutting ants have not been systematically studied in Argentina. For this reason, the aims of the present study are, using one nuclear DNA region (Tef1) and morphological information, to identify isolates of Trichoderma obtained from the nest of leafcutting ants of the genus Acromyrmex collected from different sites of Argentina and to provide new measures of diagnostic structures in order to offer information regarding the variation of the species in this country.
MATERIALS AND METHODS
Fungal Isolates.
Nests of leaf cutting ants from different sites in Argentina were sampled and their fungal gardens were collected between 2009 and 2012. We analysed eight isolates, four of them obtained from Acromyrmex lundii ant nests (T2 and T4 from two different sites at Buenos Aires, T3 from Santa Fe, and T7 from Salta), three from A. lobicornis ant nets (T1 from La Pampa, T6 from Corrientes, and T8 from Santa Fe), and one from A. aspersus ant nets (T5 from Tucumán). All of these were deposited at Laboratorio de Hormigas-UNQ collection and kept as monosporic cultures at -80º in glycerol 20% v/v for conservation.
DNA extraction.
Isolates were cultured in PDA during seven days at 25ºC and 80% RH in darkness. Genomic DNA was extracted by the CTAB method (Augustin et al., 2013). DNA was resuspended in 50 μl of Tris-EDTA Buffer (TE/10) (10 mM Tris-HCl pH 7.5; 0.1 mM EDTA). The DNA concentration was quantified using a Nanodrop 2000 (Thermo Scientific) and integrity was determined by electrophoresis using 0.8% agarose gels.
PCR amplification and sequencing.
The translation elongation factor (Tef1) was amplified using the following primers: ef-728M (5’-CACGTCGACTCCGGCAAGTC-3’), ef-2 (5’-GTGATACCACGCTCACGCTC-3’) (Samuels, 2006). The amplification was carried out in 50 μl of reaction using: 1X Taq Buffer (PBL Company), 3.5 mM MgCl2 (PBL Company), 0.2 mM of each dNTP (PBL Company), 1U Taq (Pegasus model, PBL Company), 0.4 mM for each primer (PBL Company), 25-30 ng of DNA and ddH2O to complete volume.
PCR conditions were: one minute at 94ºC, 30 cycles with one minute at 94ºC, one minute at 55ºC, one minute at 72ºC, and a final cycle with three minutes at 72°C (Barrera, 2012). PCR reactions were carried out in a Veriti 96 wells thermal cycler (Applied Biosystems). PCR products were analyzed by electrophoresis in a 1.2% agarose gel. Purification and sequencing of the PCR products were performed by Macrogen Corporation. Newly generated sequences are deposited in Gen-Bank under accession numbers (MF436982-MF436989).
Phylogenetic Analyses.
For Phylogenetic Analyses a sequences matrix was generated using the eight sequences generated in this study and those of Trichoderma obtained from the GenBank (see Supplementary appendix in the online version at http://www.ojs.darwin.edu.ar/index.php/darwiniana/article/view/724/733). Trichoderma sequences from the GenBank were selected following previous taxonomical studies (Barrera, 2012, Chaverri et al., 2015). The number of sequences per species changed according to availability of sequences in Gen-Bank. Adittionally, Hypomyces rosellus (HF911691) and Cordyceps sp. (KF226252) were used as outgroup. The result matrix was composed by 736 characters and 146 sequences (40 species), including in all cases more than one sequence per Trichoderma species. Matrix used in this study is available upon request.
Sequences were aligned using Clustal W algorithm (Gap Opening penalty = 12, Gap extension penalty = 6.66). Then were edited manually using MEGA 6 (Tamura et al., 2013). Three different analyses for phylogeny reconstruction were used: Maximum Likelihood (ML), Bayesian Inference (BI) and Maximum Parsimony (MP). Partial deletion of gaps (90 %) was used in ML, and non-deletion was done for BI. For ML and BI analysis, the program Mega 6 established the DNA sequence evolution model based on the Akaike information criterion (AIC). The model chosen was Kimura-2 parameters with a Gamma distribution to model evolutionary rate differences among sites (G parameter: 1.0128). For ML Neighbour-joining was applied to obtain the initial tree for the heuristic search. This method generated a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) method. The tree was drawn to scale, with branch lengths measured as the number of substitutions per site. Heuristic ML bootstrap analysis consisted of 1000 pseudoreplicates.
The Bayesian analysis was carried out in MRBAYES v.3.1 (Ronquist & Huelsenbeck, 2003). Two independent analyses of two parallel runs and four chains were performed with 10000000 generations and a sample frequency of 1000 trees. Bayesian Posterior probabilities (PP) were calculated using metropolis-coupled Markov chain Monte Carlo analysis until the runs (four) converged with a split frequency of 0.01. Burn-in and convergence were assessed with Tracer 1.5 (Rambaut and Drummond, 2007) and the first 25% were discarded as “burn-in”. Both runs were pooled and a consensus tree (majority rule 50%) and posterior probabilities (PP) were calculated from 15000 trees. The tree generated by this analysis was edited in the Figtree V1.4.2 software (Yrew Rambaut, Institute of Evolutionary Biology, University of Edinburgh).
Finally, the analysis was carried out using TNT 1.5 (Goloboff & Catalano, 2016), with the characters equally weighted and considering gaps as missing data and applying heuristic searches with 300 random-addition sequence replicates, with 30 random addition sequence replicates per dataset. Tree-bisection-reconnection (TBR) branch swapping was also performed. In order to obtain estimates of clade support heuristic MP bootstrap analysis consisted of 1000 pseudoreplicates was performed (TBR branch swapping) using strict consensus tree.
Maximum Likelihood topology was used to illustrate the species relationships using a three values series in nodes to show ML and MP bootstrap values and posterior probability.
Morphological identification.
All isolates were identified morphologically using macroscopic and microscopic characters. General characteristics of colony growth and phenotype were registered. Isolates were grown on PDA (Britania) during a week at 25°C in darkness. The colony area (cm2) was measured using the software ImageJ V1.47 (Wayne Rasby, National institute of Health, USA).
For microscopic studies, fungal material was mounted in water and observed under a compound microscope (Nikon, Eclipse E200). To improve visualization, Congo Red staining was used. The following microscopic characters were observed: conidiophores morphology, shape, size, and ornamentation of conidia and conidiogenous cells, plus diameter of vegetative hyphae (main axis of conidiophores). Each character was measured ten times for each isolate. To improve measurement precision, conidial size was measured using the software Micrometrics TM SE Premium, software 2.8234.
RESULTS
Phylogenetic Analysis. The matrix for Tef1 was composed by 146 taxa and 736 characters, including 61 conserved sites, 669 variable and 595 parsimony informative sites.
Phylogenetic analyses with Tef1 showed robust trees with well supported clades for each Trichoderma species (Fig. 1). The results of parsimony, likelihood and Bayesian analyses were highly concordant.
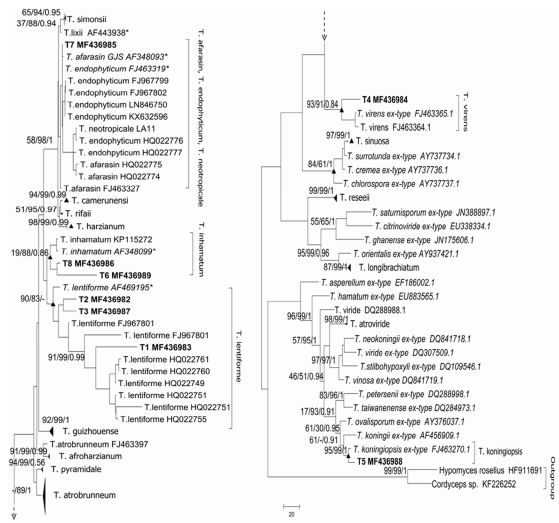
Fig. 1. Phylogeny of Trichoderma illustrating species relationships inferred from joint ML analysis of Tef1 gene analysis. The figure is divided in two subtrees. The series of three values above internal branches correspond to ML, MP and BI, respectively. Bold letters indicate our isolates. Strains corresponding to the type material are in italic letters and triangles indicate condensated nodes.
Trichoderma T1, Trichoderma T2 and Trichoderma T3 were grouped in the T. lentiforme clade (ML 90%, MP 83%, subclade T1: Ml 91%, MP 99%, BI 0.99, subclade T2-T3: ML 29%, MP 56%, BI 0.63). The sequence of Trichoderma T4 was found within the clade of the species T. virens (ML 93%, MP 91%, BI 0.84) and Trichoderma T5 was placed in the clade of the species T. koningiopsis (ML 95%, MP 99%, BI 1) (Fig. 1). Trichoderma T6 and Trichoderma T8 were grouped within the T. inhamatum clade with high supporting nodes for MP and BI (MP 88%, BI 0.86). Finally, the isolate Trichoderma T7 was placed in a clade with three species, since the molecular marker did not allowed to resolve it better (T. afarasin, T. endophyticum and T. neotropicale) (Fig. 1).
TAXONOMY
Trichoderma inhamatum Veerkamp & W. Gams. Caldasia 13: 710. 1983. Type: Colombia, Departamento Meta, Municipio de Villavicencio, “isolated from soil under maize”, 1978, O. Rangel isolated by W. Gams (isotype BPI 748209). Fig. 2.

Fig. 2. T. inhamatum. A, colony in PDA after 6 days. B-C, conidiophore. D, terminal chlamydospore. E, phialidic conidiogenous cells indicated by a black arrow. F, green conidia shown by a black arrow and terminal chlamydospore indicated with a dash arrow. Color version at http://www.ojs.darwin.edu.ar/index.php/darwiniana/article/view/724/732
Isolates exhibit the same morphology described by Veerkamp & Gams (1983) and coincide with the description made by Chaverri et al. (2015). Our isolates showed slight differences in colony and morphology of conidiogenous cells and conidia. Colonies showed a faster growing rate (30.6-43.7 cm2 after 2 days on PDA) with pustules that were absent in the original description. Conidiogenous cells bigger in our isolates (5.4-9.7 x 2.4-3.6 μm) than in the original description (4.5-8 x 2.3-3.5 μm). Conidia globose to subglobose, 2.41-3.6 x 2.24-3.67 µm.
Material examined
ARGENTINA. Corrientes. Depto. Mercedes, 2009, Gorosito isolate Trichoderma T6, “isolated from a nest of A. lobicornis Emery”. Santa Fe. Depto. San Cristóbal, 2009, Marfetán isolate Trichoderma T8, “isolated from a nest of A. lobicornis Emery”.
Trichoderma koningiopsis Samuels, C. Suárez & H.C. Evans. Studies in Mycology 56: 11. 2006. Type: Cuba, Sanctu Spiritus, Moyote Mi Ritiro, elev. 700–750 m, 21º52’ N, 80º01’ W,“on branch”, 2-VII-1993, S.M. Huhndorf 572 (BPI 802571, ex-type culture G.J.S. 93-20 = CBS 119075). Fig. 3.
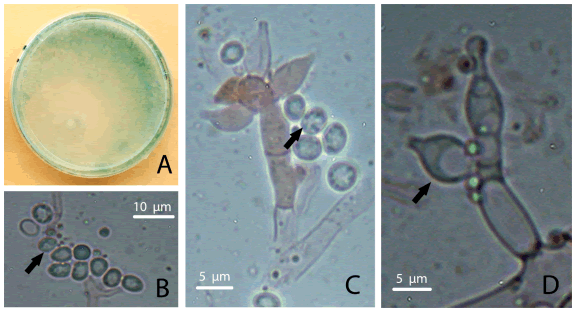
Fig. 3. T. koningiopsis. A, colony in PDA after 6 days. B-C, wrinkled dark green conidia indicated by a black arrow. D, phialidic conidiogenous cells shown by a black arrow. Color version at http://www.ojs.darwin.edu.ar/index.php/darwiniana/article/view/724/732
Isolates coincide with the original description given by Samuel et al. (2006) but show differences in conidiophore sizes, conidiogenous cells and conidia. In this study, conidiophores showed bigger main axis (5.5 µm diameter), conidiogenous cells phialidic, lageniform to ampulliform, 10-15 x 3.8-6.3 μm, and conidia were slightly bigger (4.4-6.3 x 2.5-5 μm). Chlamydospores and catenulate conidiogenous cells not seen.
Observations. The conidiogenous cells of our isolate of T. koningiopsis fall into the variation range reported in the original description (3.5-16.0 x 2.4-4.5 μm). However, the variation range showed by our isolates was wider. Moreover, T. koningiopsis isolate from Argentina obtained in previous studies also had smaller conidiogenous cells than the reported in the original description (7-10 x 2-2.5 μm) (Barrera, 2012). Additionally, the conidia diameter of our isolate was slightly bigger than the holotype description (3-6.2 x 2-3.5 μm), whereas it was similar in comparison with the description of Barrera (2012) (2-5 x 1.5-2 μm).
Material examined
ARGENTINA. Tucumán. Depto. Chicligasta, Parque de los Alisos National Park, 2012, Marfetán Isolate Trichoderma T5, “Isolated from a nest of Acromyrmex aspersus”.
Trichoderma lentiforme (Rehm) P. Chaverri, Samuels & F.B. Rocha. Mycologia 107: 558. 2015. Type: Brazil, Santa Catarina State, “on decaying leaves of Euterpe”, Aug. 1888, Ule s.n (isotype HBG #812!). Epitype: French Guiana, Commune de Saul, Mont. Galbao, base camp on NE side, near headwater of Mara River, 50–150 m, 09º10’ N, 79º50’ W, “on decaying bark possibly growing on another fungus”, 11-XI-1997, S.M. Huhndorf 3758 (BPI 744709, ex-epitype culture G.J.S. 98-6 5 CBS 100542). Fig. 4.

Fig. 4. Trichoderma lentiforme. A, colony in PDA after 6 days. B-C, conidiophores; arrows point to conidiogenous cells. D, conidia indicated by a black arrow. E-F, chlamydospores, intercalar and terminal, respectively. Color version at http://www.ojs.darwin.edu.ar/index.php/darwiniana/article/view/724/732
Isolates morphology agree with the original description published by Chaverri et al. (2015) but show some differences. In our isolates no odour was detected on PDA at 25ºC. Conidiogenous cells coincide on shape but not in size being bigger (4.3-9.3 x 2.3-5.6 μm) than previously described (5.3-5.6 x 3.5-3.5 µm). In contrast with the original description, terminal and intercalar chlamydospores were present (4.4-10.9 x 4.3-10.8 μm).
Material examined
ARGENTINA. Buenos Aires. Pdo. Berazategui, 2009, Marfetán isolate Trichoderma T2, “Isolated from a nest of A. lundii Guérin-Méneville”. La Pampa. Dpto. Lihuel Calel, Lihuel Calel National Park, 2012, Marfetán isolate Trichoderma T1, “Isolated from a nest of A. lobicornis Emery”. Santa Fe. Depto. San Cristóbal, 2009, Marfetán isolate Trichoderma T3, “Isolated from a nest of A. lundii Guérin-Méneville”.
Trichoderma virens J.H. Mill., Giddens & A.A. Foster. Arx, Beihefte zur Nova Hedwigia 87: 288, 1987. Type: Unites States of America, Indiana, Brown County, vic. Pike’s Peak, Happy Hollow Camp, 39º09’N, 86º06’W, elev. 250 m, 29-IX-1995, G.J. Samuels 95-194 (BPI 737768; CBS 109339; ATCC MYA-1298). Fig. 5.

Fig. 5. T.virens. A, colony in PDA after 6 days. B, green conidia indicated by arrows. C, conidiogenous cell pointed by black arrows. D, verticillate conidiophore highlighted by an arrow. Color version at http://www.ojs.darwin.edu.ar/index.php/darwiniana/article/view/724/732
Isolates morphology correlates with the description given by Miller et al. (1957) (as Gliocladium virens) and Chaverri et al. (2001) but show slight differences in conidiogenous cells and conidia. Conidiogenous cell ampulliform to lageniform, bigger than previous description (9-14.3 x 4-6.5 μm). Bigger conidia, subglobose, yellowish green (5.3-7.1 x 3.8-5 μm). Chlamydospores were not observed. In our isolates no pigment was detected on PDA at 25ºC.
Observations. We found bigger conidiogenous cells than in the original description (7.9-9.6 x 3.6-4.2 μm). Moreover, previously reported isolates from Argentina have similar conidiogenous cell size in comparison with our description (10-15 x 3-4 μm) (Barrera, 2012). Conversely, we found bigger conidia in comparison with the original description (4.2-4.6 x 3.6-3.8 μm) and with the description given by Barrera (2012) (4-3.5 x 3-3.5 μm).
Material examined
ARGENTINA. Buenos Aires. Depto. Mercedes, 2009, Marfetán isolate Trichoderma T4, “isolated from a nest of A. lundii Guérin-Méneville”.
Trichoderma aff. neotropicale P. Chaverri, Samuels & F.B. Rocha. Mycologia, 107: 558-590. 2015. Type: Peru, Madre de Dios, Manu, Los Amigos Biological Station, “endophytic in stems of Hevea guianensis”, 2008, R. Gazis LA11 (holotype CBS 130633, Ex-type culture G.J.S. 11-185). Fig. 6.

Fig. 6. Trichoderma sp. 7, T. aff. neotropicale. A, colony in PDA after 6 days. B, phialidic conidiogenous cells, black arrow. C, conidiophores shown by a black arrow. D, green conidia highlighted by a black arrow. Color version at http://www.ojs.darwin.edu.ar/index.php/darwiniana/article/view/724/732
Isolates agree with the original description (Chaverri et al., 2015), particularly in the morphology of the conidiogenous cells and the ramifications of the conidiophores but show wider conidiogenous cells and slight differences in conidial size. Conidiogenous cells phialidic, lageniform to ampulliform, 6-7.9 x 2.5-3.4 μm. Conidia globose to subglobose (2.9-3.5 x 2.5-3.2 μm) light green in mass, slightly smaller than original description (2.5-3.3 x 2.3-2.9 μm).
Material examined
ARGENTINA. Salta. Depto. Anta, National Park El Rey, 2012. Marfetán isolate Trichoderma T7, “isolated from a nest of A. lundii Guérin-Méneville”
DISCUSSION
This work presents the first report of T. inhamatum and T. lentiforme associated to the leaf-cutting ant nests of the genera Acromyrmex (A. lundii and A. lobicornis) and T. lentiforme is also the first reported from Argentina. There is only one report of T. lentiforme associated with leaf-cutting ants but belonging to the genus Atta (A. capiguara) from Brazil (Montoya et al., 2016). With regards to T. harzianum, T. koningiopsis and T. virens, they were previously reported associated with Atta and Acromyrmex genera from Brazil (Rodrigues et al., 2005, 2008; Silva et al., 2006; Montoya et al., 2016). In this work we confirm these results in nests from Argentina.
Molecular and morphological information allowed us to conclude that Trichoderma isolates T1, T2 and T3 belonged to Trichoderma lentiforme (Chaverri et al., 2015), however the clade of T. lentiforme have two subclades, one of them composed by T2 and T3 with low support and separated T1 (with strong support), this suggest that exist some differences among these isolates. Also, the clade of T. lentiforme is related to a more basal node with the clade of T. inhamatum, this is not surprising because Chaverri and coauthors (2015) noted this when they described the new species clarifying that is necessary to add the morphological characterization to distinguish between this two species (Fig. 1). Trichoderma T4 corresponded to Trichoderma virens (Chaverri et al., 2001), Trichoderma T5 was identified as Trichoderma koningiopsis (Samuels et al., 2006), and Trichoderma T6 and T8 belonged to Trichoderma inhamatum (Veerkamps& Gams, 1983). The isolate Trichoderma T7 was placed in the clade T. afarasin/T. endophyticum/T. neotropicale. Due to the fact that these species are cryptic for this molecular marker, we were not able to molecularly identify this isolate. However, morphologically, we found more similarities between Trichoderma T7 and T. neotropicale than with the other two species. Particularly, the morphology of the conidiogenous cells and the ramifications of the conidiophores were the diagnostic characters for the identification of the isolate.
For all isolates, morphological data agree with the original diagnosis but show some differences in the size and morphology of particular structures. The main differences were found in conidiogenous cells and conidia sizes. Moreover, isolates from Argentina obtained by Barrera (2012) have similar variation in comparison with type descriptions, suggesting that size differences are common between the original descriptions and the Argentinian isolates. However, in the case of T. virens, the variation range showed by our isolate was even wider than the variation reported by Barrera (2012). It is well known that Trichoderma is a cosmopolitan genus, so the mentioned differences in structures sizes could be associated with the place where the isolates were taken from (Chaverri et al., 2015), and therefore represent the normal morphological variation within each of the species.
The results did not show any specific association between Trichoderma and Acromyrmex species. We found T. lentiforme associated with A. lobicornis and A. lundii, but A. lobicornis was also associated with T. inhamatum. Furthermore, T. virens and T. lentiforme were isolated also from A. lundii nests. This is not surprising considering that Trichoderma species are normally found in soil and therefore ants could vector different mycoparasites to their nests. Something similar was observed in Brazil and United States of America (Montoya et al., 2016). Therefore, it seems there is no thight, co-evolving, relationship of these fungi and the leaf-cutting ants, and as such Trichoderma should be considered like an opportunistic parasite.
ACKNOWLEDGMENTS
We are thankful to Andrea Romero for her advices, Macarena Siri and Laura Velez for their kind help with the English. Financial support was provided by grants from AN P CT (PICT START UP 1936) and Programa de Investigación en Interacciones Biológicas, Universidad Nacional de Quilmes, both to P.J.F, and by a grant from UN Q (SAI2011) to J.A.M, and a fellowship from CIC to NGA; PJF and JAM thank to CONICET.
REFERENCES
1. Augustin, J. O.; J. Z. Groenewald, R. J. Nascimento, E. S. Mizubuti, R. W. Barreto, S. L. Elliot & H. C. Evans. 2013. Yet more weeds in the garden: fungal novelties from nests of leaf-cutting ants. PLoS ONE 8: 1-17. [ Links ]
2. Barrera, V. 2012. El género Hypocrea Fr. (Hypocreales, Ascomycota) en la Argentina. Estudio de la variabilidad molecular de su estado anamórfico Trichoderma. Tesis Doctoral. Facultad de Ciencias Exactas y Naturales. Universidad de Buenos Aires, P 241. [ Links ]
3. Carreiro, S. C.; F. C. Pagnocca, O. C. Bueno, M. B. Júnior, M. J. A. Hebling & O. A. da Silva. 1997. Yeasts associated with nests of the leaf-cutting ant Atta sexdens rubropilosa Forel, 1908. Antonie van Leeuwenhoek 71: 243-248. [ Links ]
4. Chaverri, P.; G. J. Samuels & E. L. Stewart. 2001. Hypocrea virens sp. nov., the teleomorph of Trichoderma virens. Mycologia 93: 1113-1124. [ Links ]
5. Chaverri, P. & G. J. Samuels. 2003. Hypocrea/Trichoderma (Ascomycota, Hypocreales, Hypocreaceae): species with green ascospores. Studies in Mycology 48: 1-116. [ Links ]
6. Chaverri, P.; F. Branco-Rocha, W. Jaklitsch, R. Gazis, T. Degenkolb & G. J. Samuels. 2015. Systematics of the Trichoderma harzianum species complex and the reidentification of commercial biocontrol strains. Mycologia 107: 14-147. [ Links ]
7. Currie, C. R.; J. A. Scott, R. C. Summerbell & D. Malloch. 1999a. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 398: 701-704. [ Links ]
8. Currie, C. R.; U. G. Mueller & D. Malloch. 1999b. The agricultural pathology of the ant fungus garden. Proceedings of the National Academy of Sciences 96: 7998-8002. [ Links ]
9. Druzhinina, I. S.; E. Shelest, & C. P. Kubicek. 2011. Novel traits of Trichoderma predicted through the analysis of its secretome. Federation of European Microbiological Societies Microbiology Letters 337: 1-9. [ Links ]
10. Fisher, P. J; D. J. Stradling, B. C. Sutton & L. E. Petrini. 1996. Microfungi in the fungus gardens of the leaf-cutting ant Atta cephalotes: a preliminary study. Mycological Research 100: 541-546. [ Links ]
11. Folgarait P.; N. Gorosito, R. Poulsen & C. R. Currie. 2011. Preliminary in vitro insights into the use of natural fungal pathogens of leaf-cutting ants as biocontrol agents. Current Microbiology 63: 250-258. [ Links ]
12. Goloboff, P. & S. Catalano. 2016. TNT, version 1.5, with a full implementation of phylogenetic morphometrics. Cladistics. DOI: 10.1111/cla.12160 [ Links ]
13. Hölldobler, B. & E. O. Wilson. 1990. The Ants. Cambridge, USA: Belknap Press of Harvard University Press, P 732. [ Links ]
14. Jaklitsch, W. M. 2009. European species of Hypocrea Part I. The green-spored species. Studies in Mycology 63: 1-91. [ Links ]
15. Little, A. E.; T. Murakami, U. G. Mueller & C. R. Currie. 2006. Defending against parasites: fungus-growing ants combine specialized behaviors and microbial symbionts to protect their fungus gardens. Biology letters 2: 12-16. [ Links ]
16. Lugo, M. A.; E. M. Crespo, M. Cafaro & L. Jofré. 2013. Hongos asociados con dos poblaciones de Acromyrmex lobicornis (Formicidae) de San Luis, Argentina. Boletín de la Sociedad Argentina de Botánica 48: 5-15. [ Links ]
17. Miller, J. H.; J. E. Giddens & A. A. Foster. 1957. A survey of the fungi of forest and cultivated soils of Georgia. Mycologia 49: 779-808. [ Links ]
18. Möller, A. 1893. Die Pilzgärten einiger südamerikanischer Ameise. Verlag Von Gustav Fischer. P 159. [ Links ]
19. Montoya, Q. V.; L. A. Meirelles, P. Chaverri & A. Rodrigues. 2016. Unraveling Trichoderma species in the attine ant environment: description of three new taxa. Antonie van Leeuwenhoek 109: 633-651. [ Links ]
20. Muchovej J.J.; T. M. Della Lucia & R. M. Muchovej. 1991. Leucoagaricus weberi sp. nov. from nest of leaf-cutting ants. Mycological Research 95:1308-1311. [ Links ]
21. Pagnocca, F. C.; M. Bacci, M. H. Fungaro, O. C. Bueno, M. J. Hebling, A. Sant’Anna & M. Capelari. 2001. RAPD analysis of the sexual state and sterile mycelium of the fungus cultivated by the leaf-cutting ant Acromyrmex hispidus fallax. Mycological Research 105: 173-176.
22. Pinto-Tomás, A. A.; M. A. Anderson, G. Suen, D. Stevenson, F. S. Chu, W. W. Cleland, P. J. Weimer & C. R. Currie. 2009. Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants. Science 326: 1120-1123. [ Links ]
23. Rambaut, A. & A. J. Drummond. 2007. Tracer 1.4. Available from http://beast.bio.ed.ac.uk/Tracer [ Links ]
24. Ribeiro, M. M. R.; K. D. Amaral & V. E. Seide. 2012. Diversity of Fungi Associated with Atta bisphaerica (Hymenoptera: Formicidae): The Activity of Aspergillus ochraceus and Beauveriabassiana. Psyche: A Journal of Entomology vol. 2012. P 6. [ Links ]
25. Rodrigues, A.; M. Jr. Bacci, U. G. Mueller, A. Ortiz & F. C. Pagnocca. 2008. Microfungal weeds in the leafcutter ant symbiosis. Microbial Ecology 56: 604-614. [ Links ]
26. Rodrigues, A.; F. C. Pagnocca, M. Bacci, M. J. A. Hebling, O. C. Bueno & L. H. Pfenning. 2005. Variability of non-mutualistic filamentous fungi associated with Atta sexdens rubropilosa nests. Folia Microbiologica 50: 421-425. [ Links ]
27. Ronquist, F. & J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572-1574. [ Links ]
28. Samuels, G. J. 2006. Trichoderma: Systematics, the Sexual State, and Ecology. Phytopathology 96: 195–206.
29. Samuels, G. J.; S. L. Dodd, B. S. Lu, O. Petrini, H. J. Schroers & I. S. Druzhinina. 2006. The Trichoderma koningii aggregate species. Studies in Mycology 56: 67-133. [ Links ]
30. Samuels, G. J. & A. Ismaiel. 2009. Trichoderma evansii and T. lieckfeldtiae: two new T. hamatum-like species. Mycologia 101: 142-156. [ Links ]
31. Scott, J. J.; K. J. Budsberg, G. Suen, D. L. Wixon, T. C. Balser & C. R. Currie. 2010. Microbial community structure of leaf-cutter ant fungus gardens and refuse dumps. PLoS One vol. 5 (3), e9922. [ Links ]
32. Silva, A.; A. Rodrigues, M. Jr. Bacci, F. C. Pagnocca & O. C. Bueno. 2006. Susceptibility of the ant-cultivated fungus Leucoagaricus gongylophorus (Agaricales: Basidiomycota) towards microfungi. Mycopathologia 162: 115-119. [ Links ]
33. Singer, R. 1986. The agaricales in modern taxonomy. 4th edition. Koleltz Scientific Books, Koenigstein. P. 918. [ Links ]
34. Tamura, K.; G. Stecher, D. Peterson, A. Filipski & S. Kumar. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725-2729. [ Links ]
35. Veerkamp, J. & W. Gams. 1983. Los hongos de Colombia VIII. Some new species of soil fungi from Colombia. Caldasia 13: 709-717. [ Links ]
36. Weber, N. A. 1972. The fungus-culturing behavior of ants. American Zoologist 12: 577-587. [ Links ]














