Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Medicina (Buenos Aires)
Print version ISSN 0025-7680On-line version ISSN 1669-9106
Medicina (B. Aires) vol.63 no.3 Buenos Aires May/June 2003
Hepatitis C virus genotypes in Cordoba, Argentina unexpected high prevalence of genotype 2
Re V.1, Lampe E.2, Fumiko Yoshida C.2, Mendes de Oliveira J.2, Lewis Ximenez L.2, Spinsanti L.1, Elbarcha O.3, Contigiani M.1
1Instituto de Virología Dr. J. M. Vanella. Facultad de Ciencias Médicas, Universidad Nacional de Córdoba;
2Fundação Oswaldo Cruz, Departamento de Virologia, Laboratório de Hepatites Virais, Rio de Janeiro, Brazil;
3Cátedra de Virología, Facultad de Ciencias Químicas, Universidad Católica de Córdoba
Abstract
To determine hepatitis C virus (HCV) genotypes circulating in the central region of Argentina, 96 consecutive anti-HCV positive subjects were studied. The presence of HCV RNA was detected in 60 samples by RT-nested PCR of the 5 noncoding region (5 NCR). Genotyping was performed by restriction fragment length polymorphism analysis of 5 NCR region combined with PCR using type-specific primers of the core region. The groups of individuals in this study included hemophilia and hemodialysis patients, injecting drug users, screened blood donors, and patients with acute or chronic liver disease, all from Córdoba, Argentina. Overall, genotype 2 was the most prevalent (55.0%), followed by genotypes 1 (38.3 %), and 3 (5.0%). Within genotype 1, subtype 1b was the most prevalent. An unexpected high prevalence of genotype 2 (61.9%) was found among patients with acute or chronic HCV infection (without known risk factors). These figures differ from other cohorts from East-Argentina where genotype 1 has been found as the most prevalent. This indicates that regional differences of genotype distribution might exist between Central and East Argentina.
Key words: Hepatitis C virus; HCV genotyping; HCV genotype 2.
Resumen
A fin de determinar los genotipos del virus de la hepatitis C (HCV) circulantes en la región central de Argentina, se estudiaron 96 individuos anti-HCV positivos. La presencia del ARN de HCV se detectó en 60 muestras mediante RT-nested PCR de la región 5 no codificante (5 NCR). La genotipificación se realizó mediante restricción enzimática y el análisis del polimorfismo de los fragmentos largos de la región 5 NCR combinada con PCR usando primers tipo específico de la región del core. El grupo de individuos estudiados incluyó pacientes hemofílicos y hemodializados, drogadictos intravenosos, donantes de sangre y pacientes con enfermedad hepática aguda y crónica, todos provenientes de Córdoba, Argentina. El genotipo 2 fue el más prevalente (55.0%), seguido por los genotipos 1 (38.3 %), con mayor prevalencia del subtipo 1b, y el genotipo 3 (55.0%). Una inesperada alta prevalencia de genotipo 2 (61.9%) se encontró entre pacientes con infección aguda o crónica por el virus HCV (sin factor de riesgo conocido). Estos hallazgos difieren de otros resultados encontrados en trabajos realizados con pacientes provenientes de la región este de Argentina, en los cuales el genotipo 1 fue encontrado como el más prevalente. Esto indica que pueden existir diferencias regionales en la distribución de genotipos HCV entre la región centro y este de Argentina.
Palabras clave: Virus de la hepatitis C; Genotipificación de HCV; HCV genotipo 2.
Hepatitis C virus (HCV) infection, which is a major cause of chronic liver disease, is strongly associated with progressive liver pathology, including cirrhosis, hepatic failure and hepatocellular carcinoma. Considering that approximately 3% of worlds population (170 million persons) are infected, chronic hepatitis C remains a serious medical problem with considerable burden on the different health care systems1.
HCV is an enveloped positive single-stranded RNA virus (9.6-kilobases) classified under the Flaviviridae family, which has remarkable genetic heterogeneity. Comparative analysis of complete or partial genomic sequences of HCV from different geographic areas identified at least six major genotypes (1-6) and a series of subtypes (a, b, c, etc)2. Although HCV genotypes 1, 2, and 3 and their subtypes have a worldwide distribution, their relative prevalence varies from one geographic area to another. In Europe and in North and South America HCV subtypes 1a and 1b have been reported as the most prevalent3, 4, 5. In Japan, subtype 1b is responsible for most of the cases of HCV infection6. While HCV subtypes 2a and 2b are relatively common in North America, Europe and Japan, subtype 2c is more commonly found in Italy7. Genotype 4 has been found mainly in Central Northern Africa (the principal genotype in Zaire and Egypt) and in the Middle East8, and types 5 and 6 have been reported in South Africa and Hong Kong, respectively9, 10.
The HCV genotype has been shown to have an important role in clinical and histological features and on the response to antiviral treatment11, 12. Genotype 1, in particular 1b, is a strong predictor for a poor response to interferon (IFN) therapy13, 14, 15. Infection with subtype 1b seems to result in a more severe liver disease leading to cirrhosis and possibly hepatocellular carcinoma11,16,17. These features suggest that subtype 1b isolates possibly represent more aggressive variants of the virus. Improved responses to IFN are found in those patients infected with either HCV genotype 2 or 3 when compared to patients infected with genotype 1 and 4. Thus, HCV genotyping is relevant not only for molecular epidemiological studies but is also potentially important in predicting the success of interferon treatment. Furthermore, the sensitivity and specificity of serological and virologic assays for detection of HCV may be influenced by the heterogeneity of HCV.
These observations have led to an overall interest in determining HCV genotypes. In Argentina, available data show that genotype 1, particularly subtype1b, is the most predominant, but a considerable percentage of non-1 genotypes have also been detected. Genotypes 2 and 3 have been reported, as well as genotypes 4 and 5 to a lesser extent18-24. For the region of Córdoba, located in the center of the country, no data on HCV genotype are available up to now.
The aim of the present study was to determine the genotypes of circulating HCV isolates in the central region of Argentina, using RFLP of the 5 NCR region combined with PCR type-specific primer of the core region.
Materials and Methods
Population: 96 consecutive anti-HCV positive patients (47 male; mean age 40.7 yr-old, range 17-70 yr and 49 female; mean age 53.4 yr-old, range 26-72 yr) were studied. All sera were collected between January 2000 and September 2001, from subjects living in Córdoba, Argentina, and referred to the Institute of Virology, School of Medicine, National University of Córdoba. The samples comprised 49 patients with sporadic hepatitis (47 chronic hepatitis and 2 acute hepatitis), 21 patients with parenteral risk of infection (9 dialysed, 7 hemophiliac, 3 intravenous drug users - IVDU - and 2 polytransfused) and 26 screened blood donors. The presence of anti-HCV antibody was determined by ELISA test III kit (Abbott, AXSYM system).
RNA Extraction: Viral RNA was extracted from 100 µl of serum with 750 µl of Trizol (Life Technologies, Rockville, MD), 1 µl (10 µg) of yeast tRNA, and 200 µl of chloroform. The mixture was mixed by pulse-vortexing for 2 min, incubated for 20 min at room temperature, and centrifuged at 14.000 rpm for 20 min. Total RNA was precipitated by isopropanol and ethanol, air dried, and then dissolved in 20 µl of diethyl pyrocarbonate treated water containing 40U of recombinant ribonuclease inhibitor (RNAsin, Promega, Madison, WI, USA).
Reverse transcription (RT) and amplification by Nested PCR: RT- Nested PCR for 5´-non coding region was used to amplify a 280 nucleotide fragment, as previously described25. First PCR was carried out with 30 cycles (denaturation at 94°C for 30 sec, annealing at 50°C for 90 sec, and elongation at 72°C for 2 min). The second PCR was done, at the same PCR conditions, with 25 cycles. PCR products were loaded onto a 1.5% agarose electrophoresis gel and visualized by staining with ethidium bromide under ultraviolet light. To avoid contamination, RNA extraction and reverse transcription, pre-PCR reagent preparation, DNA amplification, and gel electrophoresis of PCR products were performed in four separate rooms.
HCV genotyping: HCV genotyping/subtyping was deter-mined by two RT-PCR based assays, the restriction length polymorphism analysis (RFLP) of the 5´noncoding region (5´NCR), and a nested PCR with type specific primers following primary RT-PCR with consensus primers designed for the core region of HCV genome26.
RFLP: HCV genotyping by RFLP was carried out using the restriction enzymes AvaII and RsaI on PCR-amplified from 5 NCR as previously described27. Briefly, 10 µl of nested PCR products were digested separately with 5 units of restriction enzymes Rsa I and Ava II (Life Technologies). After incubation for 1.5 hs, restriction fragments were visualized on 3% agarose gel electrophoresis stained with ethidium bromide. This RFLP assay allows to distinguish between the genotypes 1, 2, 3.
Type-specific PCR assay: This was accomplished by means of the nested PCR assay originally developed by Okamoto and co-workers28 for subtyping of genotypes 1 and 2, and modified by Widell29 to identify genotype 3. All samples positive for the 5 NCR were analysed. Sequences of the HCV core region were detected after synthesis of cDNA and nested PCR with conserved primers covering positions 439 to 751 of the HCV genome26. Both, first and second PCR, were done with 35 cycles (94°C for 45 sec, 55°C for 1 min, and 72°C for 1 min 15 sec), allowing the synthesis of 313 base pair (bp) amplicons. Viral sequences amplified from the first PCR reaction with conserved primers of the core region were subjected to a second round of PCR with respective type-specific primers in two separate reactions, one for the amplification of genotypes 1a, 1b, 2a, 2b with primers type-specific for these genotypes and the other for the amplification of genotypes 1b, and 3, according to the established protocol29. Twenty five cycles of denaturation (94°C for 60 sec), annealing (63°C for 60 sec) and extention (72°C for 90 sec) were undertaken.
Statistical analysis: Ages are reported as mean (standard deviation) and were compared between the genotype groups using Students t-test. Chi-square test was used to evaluate gender distribution using STATA 6 (Stata Corporation, College Station, Texas). Statistical significance was established at P < 0.05.
Results
Among 96 patients anti-HCV positive, 60 (62.5%) were HCV RNA positive by PCR for 5 NCR. The presence of HCV RNA was detected among 42/49 (85.7%) acute or chronic hepatitis patients, in 11/21 (52.4%) of risk group and 7/26 (26.9 %) blood donors. Females tested positive more frequently than males (65.3% vs. 59.6%; P < 0.05).
The 5´ NCR was chosen for quick typing by RFLP analysis of 280 bp nested PCR product using the restriction enzymes AvaII and RsaI. By using both enzymes simultaneously it was possible to get initially information about genotypes 1, 2 and 3. In this way, type 1 was identified in 23/60 (38.3%), type 2 in 33/60 (55.0%), type 3 in 3/60 (5.0%) of the samples and one (1.7%) patient gave a indeterminate pattern.
Genotyping by using type-specific primers of the core region requires an initial amplification by PCR of genomic sequences with conserved primers followed by subtype-specific primers. Seven samples unfortunately did not have enough serum for PCR testing in core region thus, fifty three samples were submitted to PCR type-specific assay. Genotyping was successful in 39 of them. In 14 samples the HCV genotype could not be identified (all type 2 by RFLP assay), because of unsuccessful PCR amplification with the type-specific primer, and one sample gave indeterminate pattern of bands. Therefore, the genotype results among the 39 patients by PCR type-specific assay showed type 1a in 5 (12.8%), type 1b in 16 (41.0%), type 2b in 16 (41.0%), type 3 in 1 (2.6%) and indeterminate in 1 (2.6%) of patients.
The genotype distribution by RFLP or by PCR type-specific assay in the all (n=60) positive HCV RNA samples showed that 55% were identified as type 2, 38.3% as type 1, 5% as type 3, and one sample (1.7%) gave indeterminate results by both assays. Within genotype 1, subtype 1b was the most prevalent. None of the individuals was infected with more than one genotype. Table 1 shows the genotype distribution according to age and gender. A predominance (68.7%) of genotype 2 was observed in the female gender. However, this finding was not statistically significant (p=0.059). The mean age of patients infected with genotype 2 was slightly higher, but no significant association was observed between age and genotype.

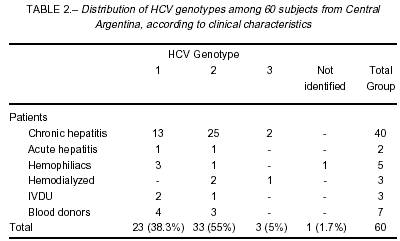
In the patients with chronic or acute hepatitis, genotype 2 was more prevalent with 26 (61.9%) out of 42 patients infected, genotype 1 accounted for 33.3% (n=14) and genotype 3 for the remaining 4.8% (n= 2) (Table 2). In our group of patients with risk factor for HCV infection (hemophiliac, hemodialyzed and IVDU; n=11) the presence of genotype 1 was identified in 5 (45.4%), genotype 2 in 4 (36.4%) and genotype 3 in 1 (9.1%) patient. One hemophiliac patient was indeterminate. Among the 7 blood donors, only genotype 1 (n=4) and 2 (n=3) were detected.
Discussion
This study shows that in the area of central Argentina, genotype 2 accounts for most of the cases of HCV infection. The genotype distribution in our population showed that 55% were infected with HCV type 2, 38% with type 1, and 5% with type 3. This high prevalence of type 2 among subjects living in Córdoba is surprising and, to our knowledge, this is the first study of the genotype distribution in this area.
These figures differ from other cohorts from Argentine patients in whom genotype 1 has been found as the most prevalent, followed by genotype 2 and 318, 19, 21, 22, 24. Quarleri et al.20 detected genotype 1 in 70.7%, genotype 2 in 21.9% and genotype 3 in 7.3% in different Argentine groups. More recently, Alfonso et al.23, reported genotype 1 in 61.4%, genotype 2 in 12.3%, genotype 3 in 23.7% and genotype 4 in 2.6% of HCV isolates from 114 infected patients. Despite the presence of genotype 2 being detected with relative high frequency in these different studies, in none of them prevalence as high as 55% has been found, as we have in the present study. However, those previous studies were probably performed on patients from the City of Buenos Aires region. This discrepancy indicates that regional differences of genotype distribution might exist between Central and East Argentina. Substantial differences in HCV genotype distribution between close geographical regions had also been observed in Taiwan. These findings implied that independent HCV outbreaks with introduction of particular HCV isolates could lead to different HCV molecular epidemiology in distinct areas and resulting in the diverse HCV genotype distribution30.
The genotype distribution of HCV infection found in our study population suggests a pattern similar to that reported in Italy, where most community-acquired cases studied so far belong to genotype 2. In a study carried out in the population of a town in Southern Italy, the HCV genotype distribution shows type 1b in 50.7%, type 2b in 0.7%, type 2c in 44.6%, type 3a in 2.7%, and type 4 in 1.3%31. Ravaggi, et al.32, also reported 54.2% (39/72 ) of genotype 2 prevalence, especially subtype 2c, in Italian patients. Similar HCV genotype distribution (around 50% of genotype 2) has been observed in other cohorts from both Southern and Northern Italy7, 33, 34.
The fact that type 2 was detected in 61.9% of patients with acute or chronic HCV infection without known risk factor highly suggests that this type may account for most of community acquired HCV in Central Argentina. Patients with risk factors for HCV infection enrolled in the present study were very few and therefore we are not allowed to reach any conclusion of genotype distribution within this group. The distribution of the two most common HCV viral types (2 and 1) was not statistically different in terms of mean age or gender and suggests that they may have had a simultaneous spread in this community.
One of the most widely used methods for genotyping HCV is a polymerase chain reaction (PCR) based on amplification of specific sequences in the core region by subtype-specific primers, as described by Widell et al.29. However, this assay, which recognizes the five most common HCV subtypes (1a, 1b, 2a, 2b and 3), was developed before the identification of new genotypes and may be inadequate to be used in some geographic areas.
Major problems were found in the identification of isolates belonging to type 2, since no genotype determination was achieved in 14 samples identified as genotype 2 by RFLP assay, because of unsuccessful PCR amplification with the type-specific primer of the core region. In the type-specific methods used, only primers for the most common subtypes, 2a and 2b, are employed, so that probably our samples could belong to another 2 subtype and could not be amplified. In fact, sequence analysis in the core region from 4 of these samples demonstrated that they belong to subtype 2c (unpublished authors data). This could explain the difference in sensitivities of the two genotyping tests used. Sequences studies on more samples are ongoing in order to better define the subtype 2 circulating in our region. The presence of subtype 2c in Argentina has been related by Quarleri et al.20, by sequence analysis in the core and NS5 regions from a sample showing discrepancies using two different methodologies; moreover, they found a strong predominance of subtype 2a/c (16/18) over 2b subtype by sequence analysis in the 5 NCR region. For detecting subtypes 2c, reported to be circulating in Italy, Spada et al.7 propose to adapt the PCR method for genotyping including primers specific to these genotypes. Taking into account the identification of subtype 2c by sequencing analysis, and recalling the great difficulty we had to identify genotype 2, it seems plausible that at least for our local strains of HCV, it might be required to use core-specific primers to subtype 2c, for correct genotype classification. Further investigation with more sequencing samples is needed to obtain the evidence that the genotype 2c isolates observed in this study are significantly different from other isolates previously characterised by sequence analysis.
In conclusion, the results reported here in point to the high prevalence of HCV genotypes 2 in patients living in the Central Region of Argentina, and most importantly that these findings have consequential therapeutic implications in view that infections with HCV 2 or 3 were independent predictors for sustained response to IFN monotherapy15,35 or IFN/ribavirin combination11,36. In general, patients with the genotype 1 infection should receive 48 weeks of therapy, and those with genotypes 2 or 3 infection only 24 weeks. Viral load estimations are problematic because of normal fluctuations (up to 0.5-10 log), assay variability and lack of a universally accepted standard; thus, viral load testing is not recommended routinely at present37. Furthermore, the determination of HCV genotypes present in different regions of Argentina is very important for epidemiological surveillance, blood-donor screening and adequacy of foreign serological and genotyping tests.
Acknowledgements: This study was supported in part by grants from CAPES-cordenação de Aperfeiçoamento de Pessoal de Nível Superior (Brazil), and SeTCIP-Secretaría para la Tecnología, la Ciencia y la Innovación Productiva (Argentina).
1. World Health Organization (WHO). Wkly Epidemiol Rec 1999; 49: 421-8. [ Links ]
2. Simmonds P, Alberti A, Alter H, et al. A proposed nomenclature of hepatitis C viral genotypes. Hepatology 1994; 19: 1321-4. [ Links ]
3. Zein NN, Rakela J, Krawitt EL, Reddy KR, Tominago T, Persing DH and the Collaborative Study Group. Hepatitis C virus genotypes in the United States: epidemiology, pathogenicity and response to interferon therapy. Ann Intern Med 1996; 125: 634-9. [ Links ]
4. McOmish F, Yap PL, Dow BC, et al. Geographical distribution of hepatitis C virus genotypes in blood donors: an international collaborative survey. J Clin Microbiol 1994; 32: 884-92. [ Links ]
5. Nousbaum J, Pol S, Nalpas B, Landais P, Berthelot P Bréchot C and the Collaborative Study Group. Hepatitis C type 1b (II) infection in France and Italy. Ann Intern Med 1995; 122: 161-8. [ Links ]
6. Takada N, Takase S, Takada A, Date T. Differences in the hepatitis C genotypes in different countries. J Hepatol 1993; 17: 277-83. [ Links ]
7. Spada E, Ciccaglione AR, Dettori S, et al. Genotyping HCV isolates from Italy by type–specific PCR assay in the core region. Res Virol 1998; 149: 209-18. [ Links ]
8. Chamberlain RW, Adams N, Saeed AA, Simmonds P, Elliott RM. Complete nucleotide sequence of a type 4 hepatitis C virus variant, the predominant genotype in the Middle East. J. Gen. Virol 1997; 78:1341-7. [ Links ]
9. Simmonds P. Viral heterogeneity of the hepatitis C virus. J Hepatol 1999; 31: 54-60. [ Links ]
10. Zein N. Clinical significance of hepatitis C virus genotypes. Clin Microbiol Rev 2000; 13: 223-35. [ Links ]
11. Poynard T, Marcellin P, Lee SS, et al. Randomized trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. International Hepatitis Interventional Therapy Group. Lancet 1998; 31; 352:1426-32. [ Links ]
12. Nagayama K, Kurosaki M, Enomoto N, Miyasaka Y, Marumo F, Sato C. Characteristics of hepatitis C viral genome associated with disease progression. Hepatology 2000; 31: 745-50. [ Links ]
13. Martinot-Peignoux M, Marcellin P, Pouteau M, et al. Pretreatment serum hepatitis C virus RNA levels and hepatitis C virus genotype are the main and independent prognostic factors of sustained response to interferon alfa therapy in chronic hepatitis C. Hepatology 1995; 22: 1050-6. [ Links ]
14. Boyer N, Marcellin P. Pathogenesis, diagnosis and mana-gement of hepatitis C. J Hepatology 2000; 32: 98-112. [ Links ]
15. Zeuzem S, Herrman E, Lee J-H, et al. Viral Kinetics in patients with chronic hepatitis C treated with standard or peginterferon alfa 2a. Gastroenterology 2001; 120: 1438-47. [ Links ]
16. Silini E, Bottelli R, Asti M, et al. Hepatitis C virus genotypes and risk of hepatocellular carcinoma in cirrhosis: a case-control study. Gastroenterology 1996; 111: 199-205. [ Links ]
17. Bruno S, Silini E, Crosignani A, et al. Hepatitis C virus genotypes and risk of hepatocellular carcinoma in cirrhosis: a prospective study. Hepatology 1997; 25: 754-8. [ Links ]
18. Oubiña JR, Quarleri JF, Rudzinski M, Parks C, Badía I, Gonzalez Cappa SM. Genomic characterization of hepatitis C virus isolates from Argentina. J Med Virol 1995; 47: 97-104. [ Links ]
19. Picchio GR, Nakatsuno M, Boggiano C, et al. Hepatitis C (HCV) genotype and viral titter distribution among Argentinean haemophilic patients in the presence or absence of human immunodeficiency virus (HIV) co-infection. J Med Virol 1997; 52: 219-25. [ Links ]
20. Quarleri JF, Robertson BH, Mathet V, et al. Genomic and phylogenetic analysis of hepatitis C virus strains from Argentina. Medicina (Buenos Aires) 1998; 58: 153-9. [ Links ]
21. Findor J, Sorda J, Daruich J, et al. Distribución de geno-tipos del virus de la hepatitis C en una población Argentina de drogadictos endovenosos. Medicina (Buenos Aires) 1999; 59: 49-54. [ Links ]
22. Oubiña JR, Quarleri JF, Sawicki MA, et al. Hepatitis C Virus and GBV-C/Hepatitis G virus in Argentine patients with porphyria cutanea tarda. Intervirology 2001; 44: 215-18. [ Links ]
23. Alfonso V, Flichman D, Sookoian S, Mbayed A, Campos R. Phylogenetic characterization of genotype 4 hepatitis C virus isolates from Argentina. J Clin Microbiol 2001; 39: 1989-92. [ Links ]
24. Gismondi MI, Preciado MV, Badia I, Ferro A, Galoppo C, Grinstein S. Genotype characterization of hepatitis C virus infection in children. Medicina (Buenos Aires) 2001; 61: 815-20. [ Links ]
25. Mendes de Oliveira J, Rispeter K, Viazov S, Saback F, Roggendorf M, Yoshida C. Differences in HCV antibody patterns in haemodialysis patients infected with the same virus isolate. J Med Virol 2001; 63: 265-70. [ Links ]
26. Viazov S, Kuzin S, Paladi N, et al. Hepatitis C virus genotypes in different regions of the former Soviet Union (Russia, Belarus, Moldova and Uzbekistan). J Med Virol 1997; 53: 36-40. [ Links ]
27. Driesel G, Wirth D, Stark K, Baumgarten R, Sucker U, Schreier E. Hepatitis C virus (HCV) genotype distribution in German isolates: studies on the sequence variability in the E2 and NS5 region. Arch Virol 1994; 139: 379-88. [ Links ]
28. Okamoto H, Okada S, Sugiyama Y, et al. Nucleotide sequence of the genomic RNA of hepatitis C virus isolated from a human carrier: comparison with reported isolates for conserved and divergent regions. J Gen Virol 1991; 72: 2697-704. [ Links ]
29. Widell A, Shev S, Mansson S, et al. Genotyping of hepatitis C virus isolates by a modified polymerase chain reaction assay using type specific primers: epidemio-logical applications. J Med Virol 1994; 44: 272-9. [ Links ]
30. Yu M-L, Chuang W-L, Chen S-C, et al. Changing prevalence of hepatitis C virus genotypes: Molecular epidemiology and clinical implications in the hepatitis C virus Hyperendemic areas and a tertiary referral center in Taiwan. J Med Virol 2001; 65: 58-65. [ Links ]
31. Guadagnino V, Stroffolini T, Rapicetta M, et al. Prevalence, risk factors, and genotype distribution of hepatitis C virus infection in the general population: a community-based survey in southern Italy. Hepatology 1997; 26: 1006-11. [ Links ]
32. Ravaggi, A., Zonaro A., Marian M.G., Puoti M., Albertini A., Cariani E. Distribution of viral genotypes in Italy deter-mined by hepatitis C virus typing by DNA immunoassay. J Clin Microbiol 1994; 32: 2280-4. [ Links ]
33. Maggi G, Armitano S, Brambilla L, et al. Hepatitis C infection in an Italian population not selected for risk factors. Liver 1999;19: 427-31. [ Links ]
34. Osella A, Giovanni M, Guerra V, et al. Hepatitis C virus genotypes and risk of cirrhosis in Southern Italy. Clin Infect Dis 2001; 33: 70-5. [ Links ]
35. Kohara M, Tanaka T, Tsukiyama-Kohara K, et al. Hepatitis C virus genotypes 1 and 2 respond to interferon-alpha with different virologic kinetics. J Infect Dis 1995; 172: 934-8. [ Links ]
36. Bjoro K, Bell H, Hellum KB, et al. Effect of combined interferon-alpha induction therapy and ribavirin on chronic hepatitis C virus infection: a randomized multicentre study. Scand J Gastroenterol 2002; 37: 226-32. [ Links ]
37. Mc Hutchison JG. Hepatitis C advances in antiviral therapy: What is accept treatment now? J Gastroenterol Hepatol 2002; 17: 431-4 [ Links ]
Recibido: 26 de diciembre de 2002
Aceptado: 21 de abril de 2003














