Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Medicina (Buenos Aires)
versión impresa ISSN 0025-7680versión On-line ISSN 1669-9106
Medicina (B. Aires) v.66 n.4 Buenos Aires jul./ago. 2006
Hiv-1 genetic diversity in Argentina and early diagnosis of perinatal infection
Paula C. Aulicino1, Manuel Gómez Carrillo2, Julieta Kopka1, Andrea M. Mangano1, Marcelo Ovejero1, Luisa Sen*1
1Laboratorio de Biología Celular y Retrovirus, Hospital de Pediatría Juan P. Garrahan;
2Centro Nacional de Referencia para el SIDA, Buenos Aires
Postal address: Dra. Luisa Sen, Laboratorio de Biología Celular y Retrovirus, Hospital de Pediatría J.P. Garrahan, Combate de los Pozos 1881, 1245 Buenos Aires, Argentina. Fax (54-11) 4308-5325, E-mail: lsen@garrahan.gov.ar
Abstract
HIV-1 diagnosis of perinatally exposed children is usually performed by molecular biology-based methods, allowing the direct detection of the virus. Thus, HIV-1 genomic variability within and across strains plays a major role in relation to the sensitivity of these tests, often leading to misdiagnosis. We describe the performance of an in-house multiplex nested PCR (nPCR) for early detection of HIV-1 infection in perinatally exposed children born in Argentina, where the percentage of diverse BF recombinants is as high as 80%. After evaluation of 1316 HIV-1 perinatally exposed children collected over a 7-year period, the specificity and sensitivity of the diagnostic nPCR was of 100% and 99.2% respectively, with only two false negative cases indicating a good performance of the diagnostic nPCR in the Argentine pediatric cohort. In search of unusual HIV-1 subtypes among 22 HIV-1 infected cases presenting partial or complete HIV-1 gene amplification failure, we performed phylogenetic and recombination analysis of a vpu-env fragment in addition to gag and env Heteroduplex Mobility Assay screening. The most unusual findings included two subtypes A and a novel BC recombinant, while the majority of the strains were a variety of different BF recombinants. These results indicate the presence of novel and heterogeneous genotypes in our country and the need of continuous viral surveillance not only for diagnostic test optimization but also for the eventual implementation of a successful vaccine.
Key words: HIV-1 epidemiology, BF Recombinants, HIV-1 subtypes, Perinatal transmission, HIV-1 diagnosis
Resumen
La diversidad genética del HIV-1 en la Argentina y el diagnóstico temprano de la infección perinatal. El diagnóstico temprano de infección por HIV-1 en niños expuestos perinatalmente al virus se realiza con técnicas de biología molecular, detectando el virus en sangre. Por ello, la variabilidad genómica intra e inter subtipo del HIV-1 juega un rol importante en relación a la sensibilidad de estos tests. Describimos aquí la performance de una PCR multiplex anidada artesanal (nPCR), rutinariamente usada para el diagnóstico temprano de la infección por HIV-1 en niños expuestos por vía perinatal en Argentina, donde cerca del 80% de las cepas circulantes son diversas recombinantes BF. La nPCR se utilizó para el diagnóstico de infección en 1316 niños en un período de 7 años, obteniéndose una especificidad de 100% y una sensibilidad de 99.2%, con sólo 2 falsos negativos. En busca de subtipos inusuales de HIV-1 entre los 22 casos que presentaron falla completa o parcial en la amplificación por nPCR, se subtipificaron fragmentos de gag y env mediante el Ensayo de Movilidad de Heteroduplex, y de los genes vpu-env por análisis filogenético y de recombinación. Se encontraron un subtipo A en 2 hermanos, y una recombinante BC, mientras que el resto de las cepas de HIV-1 pertenecieron a diversas recombinantes BF. Los resultados indican que en nuestro país circula una variedad de genotipos de HIV-1 con influencia sobre el diagnóstico. El continuo monitoreo de las cepas circulantes es necesario tanto para la optimización de las técnicas de diagnóstico como para la implementación de una vacuna efectiva en el futuro.
Palabras clave: Epidemiología molecular, Recombinantes BF, Subtipos de HIV-1, Transmisión vertical, Diagnóstico de HIV-1.
Early diagnosis of HIV-1 infection in infants born to seropositive women is relevant for good medical management of the newborn and prompt initiation of anti-retroviral therapy, as well as for proper family counseling and assistance.
Children born to HIV antibody-positive women passively acquire maternal anti-HIV antibodies, which usually persist up to 7 to 12 months of age. Therefore, the use of assays for direct detection of the virus is necessary for early stage diagnosis of HIV-1 infection in children under 18 months of age. The diagnostic method used is a DNA polymerase chain reaction (PCR) with high sensitivity and specificity, relative low cost, easy implementation and rapid turnaround time compared to peripheral blood mononuclear cells (PBMCs) culture for viral isolation or plasma HIV-1 p24 antigen detection1. Commercially available PCR-based tests which detect a single HIV-1 gag gene fragment have been thoroughly tested in the United States and Europe, where HIV-1 subtype B is the predominant subtype2, and despite the modifications introduced to improve detection of all circulating HIV-1 strains, subtype-specific detection problems for non-B HIV-1 genotypes still persist, mainly because of the emergence of recombinant forms and unusual subtypes3-5. As non-B subtypes usually circulate in developing countries, where commercial tests are not easily available, in order to reduce costs and improve the detection of local circulating strains, "in-house" PCRs are often developed6-8.
In Argentina, nearly 80% of HIV-1 vertically transmitted strains are BF recombinants, and most of the remaining 20% are subtype B9. Genomic analysis of the diverse BF recombinants in current circulation showed that they are mainly subtype F, with short subtype B fragments usually intercalated at the long terminal repeats (LTRs) and genomic regions encoding for reverse transcriptase (RT), protease (PR) and VPU proteins10, 11. The BF recombinants seem to be restricted to Latin America, with the highest prevalence reported in Buenos Aires, Argentina12. However, information on reactivity to BF recombinants by commercial or "in-house" PCR-based diagnostic methods is not available. Thus, our aim was to evaluate the performance of an "in-house" multiplex nested PCR (nPCR) for early diagnosis of HIV-1 infection in a cohort of Argentine infants, where BF recombinants predominate. Among the samples with failure to amplify certain viral fragments, HIV-1 subtype was investigated to determine if unusual subtypes may have impaired PCR amplification.
Materials and Methods
Study subjects
The study retrospectively included 1316 children born to HIV-1 infected mothers and referred to the Hospital de Pediatría "J.P. Garrahan" between 1997 and 2002 for diagnosis of perinatal HIV-1 infection. Only children with two nPCRs prior to 18 months of age and with HIV-1 serological confirmation by Enzyme immunoassay (EIA) and Western Blot (WB) after turning 18 months were included in the study. Samples from 744 children were obtained at our Hospital, and samples from 572 infants were delivered to our Laboratory from the following institutions: Hospital Zonal General de Agudos Dr. Ramón Carrillo, Los Polvorines (n= 12), Hospital Materno Infantil Ramón Sardá (n= 281) Hospital Provincial Neuquén (n= 65), Policlínico Regional J.D. Perón - Villa Mercedes- San Luis (n= 9), Hospital Central de Mendoza (n= 42), Hospital General de Agudos Dr. Carlos G. Durand (n= 78), Hospital de Agudos Ramos Mejía (n= 70) and Hospital General de Agudos Parmenio Piñero (n= 15). Informed consent was obtained from parents or legal guardians in all cases.
Specimen collection
Whole blood specimens were obtained by venipuncture into EDTA-containing tubes and processed at the Laboratory of Cellular Biology and Retroviruses, Hospital de Pediatría J.P. Garrahan. PBMCs were purified by Ficoll Hypaque density gradient centrifugation as previously described13, and used for HIV-1 DNA testing and HIV-1 subtyping. Plasma samples were stored at -70°C for viral load and/or p24 measurements. When cell recovery was higher than 5x106 PBMCs, HIV-1 isolation by coculture was performed.
Plasma HIV-1 viral load and p24 determination
HIV-1 plasma RNA copies were measured by HIV-1 RNA QT Nuclisens (Organon Teknika, Boxtel, The Netherlands) or Amplicor HIV-1 Monitor test v 1.5 (Roche Diagnostic Systems, Branchburg, U.S.A.) according to the manufacturer's instructions. HIV-1 p24 core antigen (p24 Ag) was measured with a commercial ELISA (VIRONOSTIKA HIV-1 Antigen, Biomérieux, France).
PBMC coculture
HIV-1 was isolated by means of cell cocultures as previously described14. Briefly, 3-6 x 106 PBMCs from the patients were cocultured with an equal amount of PBMCs obtained from HIV-seronegative blood donors, prestimulated with phytohe-maglutinin (PHA), in IL-2- containing medium. The cultures were maintained for over a month, and cell-free supernatants were collected on days 2 to 5 and twice a week thereafter. Every week, fresh PHA-stimulated donor cells were added to the cultures. Viral production was determined by measuring p24 HIV-1 core antigen in PBMC culture supernatants with a commercial ELISA (HIVAg 1 Monoclonal Abbott Laboratories, USA). Positivity was considered when p24 Ag production was higher than 50 pg/ml at any point during the month of coculture.
Multiplex nested PCR for early diagnosis of HIV-1 infection
The nPCR was developed in 199615 for the simultaneous amplification of a 132 bp gag and a 372 bp env HIV-1 gene fragments and a 524bp human beta-actin fragment (internal control) in a two-round nested PCR.
DNA was amplified in 50-µl reaction mixtures containing 1x Taq Buffer, 0.2 mM dNTPs each, 0.2 µM of each primer, 1.5 U of Taq polymerase and 5 mM MgCl2 in the first round or 2.3 mM MgCl2 in the second. In the initial PCR, 15 µl of the genomic DNA (corresponding to 1.5 x 105 cells) was used as an input, and 5 µl of the 1st round reaction mixture was used for the 2nd round. Previously described primer pairs16 JA9 (env, forward), JA12 (env, reverse), JA4 (gag, forward), JA7 (gag, reverse), together with b-actin primers b-1 (forward) and b-2 (reverse) were used for the 1st round amplification cycle and JA10 (env, forward), JA11 (env, reverse), JA5 (gag, forward) and JA6 (gag, reverse) were used for the 2nd round. For both rounds, DNA was denatured for a 5-min period at 95°C followed by 24 amplification cycles of 30 sec denaturation at 95°C, 30 sec annealing at 50°C, and 30 sec extension at 72°C plus 2 sec per cycle, and a subsequent extra 5 min extension step at 72°C followed by soaking at 15°C. b-actin primers´ sequences were as follows: b-1: 5´GGA CCT GAC TGA CTA CCT CAT GAA; b-2: 5´GAT CCA CAT CTG CTG GAA GGT CG. Second round PCR products were resolved on 2% agarose gels stained with ethidium bromide and observed under UV light.
Infant HIV-1 infection status
HIV-1 infection was confirmed when at least one of the two HIV-1 gene fragments (132 bp gag and/ or 372 bp env) could be amplified in 2 consecutive beta-actin- positive blood samples. In case of discordant nPCR results, a third sample was collected and tested. All nPCR results were serologically confirmed by EIA (VIDAS HIV DUO, Biomerieux Argentina or HIV-1/ HIV-2 III Plus IMX, Abbot Argentina or Determine HIV-1/2 Abbot Argentina) and WB (New Lav Blot I, BioRad France) at or after 18 months of age.
HIV-1 genotyping
Both gag and env HIV-1 gene fragments were subtyped by the Heteroduplex Mobility Assay (HMA) as previously described17, 18 using the HIV-1 env and gag Genetic Subtyping Kits kindly provided by the AIDS Research and Reference Reagent Program (Division of AIDS, National Institute of Allergy and Infectious Diseases [NIAID], National Institutes of Health [NIH]). Briefly, a nPCR with two 35 primer-extension cycles was used to amplify a 1.25 Kb and then a 0.7 Kb internal fragment of the env HIV-1 gene (encoding the V3-V5 region of the envelope protein) with primers ED5/ ED12 for the first round and ES7/ ES8 for the second round; the 460 bp gag gene fragments were amplified with primers H1G777/ H1P202 for the 1st round and Gag1584/ g17 for the 2nd round. Heteroduplexes were formed by mixing the 2nd round product of DNA from each patient with subtype specific references and resolved in a 5% polyacrilamide gel with 20% urea for gag fragments and without urea for the env fragments in Tris-Borate-EDTA for 2 hours at 250 volts. Gels were stained with ethidium bromide and observed under UV light.
For identification of BF recombinants, a nearly 600 bp vpu-env HIV-1 fragment containing a common breakpoint for most Argentine BF recombinants including CRF12_BF, was amplified by nested PCR as described by Gómez Carrillo et al9. Vpu-env PCR products were sequenced by the dideoxy method using the DYEnamic ET Terminator Cycle sequencing kit (Amersham Biosciences, England). All sequencing reactions were run in an ABI PRISM 310 automated sequencer and assembled with the DNA Sequencing Analysis Software v 3.3 (Applied Biosystems, USA). Sequences were submitted to Genbank under accession numbers AY284977 to AY284993.
A multiple alignment of vpu-env sequences was made with HIV-1 reference sequences: subtype A (SE7253 from Somalia and SE7535 from Uganda), subtype B (MN, WR27 from USA and RL42 from China), subtype C (BW15 and BW16 from Botswana), subtype F (BR020 from Brazil, F9363 from Finland and VI850 from Democratic Republic of Congo) and BF recombinants (ARMA159 and ARMA185 from Argentina) using ClustalX19. Alignments were visually corrected with BioEdit version 5.0.920. A phylogenetic tree was constructed by Neighbor-Joining using the Kimura 2-parameter model with the MEGA version 2.1 program21. Bootstrap analyses were done to assess the stability of the nodes. Recombination breakpoints within the vpu-env region were evidenced by bootscan analysis with Simplot version 2.5 based on 100 resamplings, supporting branching with the reference sequences within a 200 bp window moving in steps of 20 bases.
Results
Sensitivity and specificity of the multiplex nested PCR used for HIV-1 early diagnosis
Of the 1316 HIV-1 perinatally exposed children studied, 241 acquired HIV-1 infection from their mothers, confirmed by the presence of anti-HIV-1 antibodies after 18 months of age (detected by EIA and WB). Of them, 239 were also positive by nPCR, amplifying gag and/or env HIV-1 fragments together with b-actin from at least two different blood samples, whereas in 2 cases both viral fragments persistently failed to amplify, rendering false negative nPCR results. In the 1075 infants seronegative for HIV-1, none of the HIV-1 gene fragments were amplified by nPCR. Thus, nPCR rendered 99.2% sensitivity and 100% specificity.
We looked for unusual subtypes in the 2 false negative cases and also among the 20 cases with persistent failure to amplify one and the same viral fragments (gag or env) on different samples. Information of the 22 cases with amplification problems, divided into 3 groups, are detailed in Table I. Group 1 includes the 2 false negative nPCR cases. Group 2 includes 14 infected infants with repeated amplification only for gag HIV-1 fragment by nPCR, and group 3 includes 6 cases positive only for the HIV-1 env fragment. PA-21 represents an exception within group 2, as he was nPCR- positive for the gag fragment in a single sample, but on subsequent blood samples no HIV-1 amplification was achieved. However, HIV-1 isolation from PBMC coculture and detection of HIV-p24 antigen in plasma (data not shown) at 3 months of age allowed the diagnosis of HIV-1 infection in PA-21 prior to serological confirmation. Table I also includes information about date of birth, sex, breast-feeding time length, and infant age at nPCR-testing, together with plasma viral loads (VL) and PBMC coculture data obtained from the same collected blood samples. HIV-1 p24 antigen was also detected in plasma samples from infants PA-24 and PA-26 at 5 and 6 months of age respectively. High plasma viral loads, HIV-1 isolation from PBMC cocultures and/or presence of p24 antigen in plasma confirmed an ongoing HIV-1 infection at the time of nPCR- testing in most cases studied, despite showing conflicting nPCR results.
TABLE 1.- HIV-1 studies and clinical data from 22 infected children with gag and/or env amplification failure by the diagnostic nPCR.
TABLE 1.- Contiinuation
HIV-1 gag and env subtyping by HMA
Gag and env-HMA genotypic characterization of the 22 HIV-1 samples with amplification failures is detailed in Table 1. The first HIV-1 false negative case detected was patient PA-11. Her older brother had been previously diagnosed HIV-1 infected at our center in 1997 with some initial diagnostic problems (PA-21 located in Group 2). HMA genotyping revealed that both siblings and their mother, an immigrant from Peru, carried a subtype A HIV-1 strain. The second false negative case, PA-12, was env F/gag F by HMA.
Of the 14 cases with nPCR amplification for the gag segment alone (group 2), by HMA 10 were gag F/ env F, 1 gag B/ env B, 1 gag A (PA-21) and 2 gag F. Of the 6 cases with nPCR amplification for the env segment alone (group 3), 3 were gag F/ env F,1 gag B/ env F, 1 gag F/ env B and 1 gag B / env C (PA-33). The latter was confirmed by sequencing and phylogenetic analysis of the fragments from samples drawn at two different time points (data not shown)22, 23.
Phylogenetic analysis of the vpu-env segment
To further characterize the HIV-1 genotype of the 22 cases described in Table I, a 600 bp fragment of the HIV-1 genome comprising the vpu gene and the 5´portion of env was sequenced for recombination and phylogenetic analysis.
Phylogenetic analysis showed that all the cases initially subtyped as F by gag and/or env HMA were BF recombinants clustering away from F reference subtypes and between the clusters defined for F and B subtypes (Fig 1a). Unfortunately, the vpu-env fragments could not be amplified for isolates ARA616 and ARA836. Isolate ARE195 from PA-33, classified as gag C/ env B by HMA, clustered together with subtype B references at vpu-env (Fig 1a). Isolate ARC315 from PA-11 clearly clustered with subtype A references, confirming HMA results, while the vpu-env fragment of her brother PA-21 could not be amplified for sequencing.
In search for BF recombination breakpoints, vpu-env sequences were analyzed by bootscanning with SimPlot. High bootstrap values confirmed a common 5´B/ 3´F mosaic pattern in most of the isolates initially genotyped as F or BF by HMA (Fig 1b). The analysis also allowed the identification, within the HIV-1 vpu-env region, of dissimilar BF recombinant structures, like the ones found in isolate ARC156 from patient PA-32 (Fig 1c) included in Group 3, and in isolate ARB717 from patient PA-12 (Fig 1d) included in Group 1 for rendering a false negative nPCR result. ARC156 showed an inverted 5´F/3´B vpu-env sequence, and ARB717 showed the presence of 3 recombination breakpoints within the fragment.
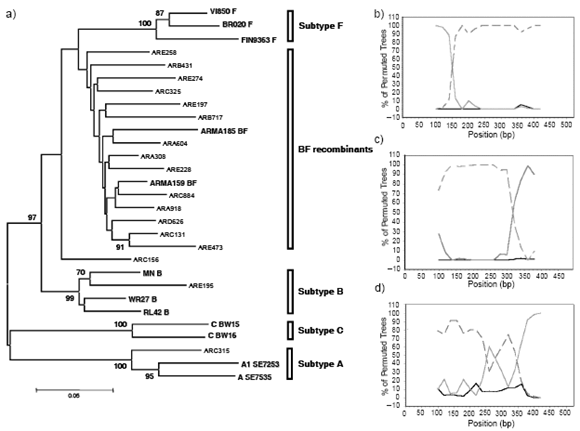
Fig. 1.- (a) Neighbour- joining phylogenetic analysis of the 600bp vpu-env sequences from HIV-1 isolates detailed in Table 1 compared to subtype B, F, C, A and BF references. (b) Representative bootscan analysis for most 600pb vpu-env sequences. (c) Bootscan analysis of ARC156 vpu-env sequence (d) Bootscan analysis of ARB717 vpu-env sequence. The horizontal axis represents nucleotide distance of the midpoint of the window from the 5´end of the query sequence. The vertical axis represent the percentage of trees (using 100 bootstrap replicates) that support branching with the consensus reference sequences: subtype B (continuous grey) and subtype F (dashed gray). Subtype D (continuous black) was used as outgroup.
To determine if the distribution of HIV-1 subtypes may differ between the groups with and without amplification failure by nPCR, we further performed vpu-env sequencing on 23 gag and env nPCR positive cases from the same cohort of HIV-1 vertically infected Argentine children. We found 18 BF recombinants (78.3%) and 5 subtype B strains (21.7%), with absence of other subtypes.
Discussion
The detection of HIV-1 proviral DNA by means of an "in-house" multiplex nested PCR has shown to be a very valuable tool for the diagnosis of HIV-1 infection in our pediatric population where BF recombinants predominate. The diagnostic assay showed a high sensitivity and specificity due to the combined amplification of gag and env HIV-1 fragments. However, diversity in both HIV fragments, within and across clades, resulted in repeated lack of detection of one (8%) or both HIV-1 fragments (0.8%) tested by nPCR in HIV-1 infected infants, thus alerting the presence of unusual or different HIV-1 strains in circulation among the Argentine pediatric population.
Most commercial HIV-1 early diagnostic tests detect only the gag gene fragment. Our HIV-1 nPCR assay included the amplification of an additional env gene fragment, increasing the sensitivity from 96.7% (gag alone) to 99.2% (gag plus env). Thus, the simultaneous PCR amplification of 2 viral fragments in our cohort can improve the chances of HIV-1 detection and also reduce costs.
Among the specimens studied with repeated partial or complete lack of amplification by nPCR, 13% corresponded to very unusual HIV-1 subtypes in Argentina. PA-33, born very near the Brazilian frontier, carried a novel HIV-1 BC recombinant22, 23 and the subtype A found in the Argentine siblings PA-11 and PA-21 was acquired through vertical transmission from their Peruvian mother. Interestingly, subtype A was only reported in 0.1% of the circulating strains in all Latin-America and in only one out of 224 cases studied in Peru24, 25. In comparison, our control group with gag/env positive by nPCR, only B and BF genotypes were found at the vpu-env gene fragment. Through this study, we also confirmed the rarity of pure subtype F in Argentina10-13. Overall, we found that new HIV-1 subtypes and diverse recombinant forms could reduce the sensitivity of the diagnostic method applied in children. Nevertheless, the sensitivity remained as high as 99.2%, due to the detection of two different HIV-1 genomic regions and the criteria used for HIV-1 infection. Although initially restricted to Latin-America, BF recombinants have been recently detected in European countries like Italy and Spain26. Thus, it will be necessary to include BF recombinants into the international HIV-1 reference and Standard Panels for validation of diagnostic and follow-up tests used worldwide. Continuous surveillance for detection of emerging variants should be carried out not only for a regular update and optimization of HIV-1 molecular diagnosis but also for the future implementation of effective vaccines.
Acknowledgements: Sponsorship: The work was supported in part by the Agencia Nacional de Promoción Científica y Tecnológica Award N° 990506326, Ramón Carrillo - Arturo Oñativia funds and CONICET PIP N° 1052/98.
The authors gratefully thank the participants and all the coordinators affiliated with this study. To A Sarubi and M Ortiz de Zárate from Hospital Materno Infantil Ramon Sardá; M Pasaresi, C Terrones and S Hermosid from Hospital Carlos Durand; L Lima and V Bittar from Hospital Central de Mendoza; M Iacono and R Micci from Hospital Provincial Neuquén; R Zlatkes from Hospital Ramos Mejía; M Rabal and G Nava from Policlínico Regional "Perón"- Villa Mercedes-; S Brotzmann and A Collia from Hospital Ramon Carrillo and M Luis from Hospital Piñero. We also wish to thank Marcelo Batalla, Carmen Gálvez and Andrés Peralta for technical assistance and also Carlos Rocco, Elliot Cowan and Sergio Rosenzweig for helpful discussion and critical review of the manuscript.
References
1. Kovacs A, Xu J, Rasheed S, et al. Comparison of a rapid nonisotopic polymerase chain reaction assay with four commonly used methods for the early diagnosis of human immunodeficiency virus type 1 infection in neonates and children. Pediatr Infect Dis J 1995; 14: 948-54.
2. Cassol S, Butcher A, Kinard S, et al. Rapid screening for early detection of mother-to-child transmission of human immunodeficiency virus type 1. J Clin Microbiol 1994; 32: 2641-5.
3. Bogh M, Machuca R, Gerstoft J, et al. Subtype-specific problems with qualitative Amplicor HIV-1 DNA PCR test. J Clin Virol 2001; 20: 149-53.
4. Loussert-Ajaka I, Descamps D, Simon F, Brun-Vezinet F, Ekwalanga M, Saragosti S. Genetic diversity and HIV detection by polymerase chain reaction. Lancet 1995; 346: 912-3.
5. Barlow KL, Tosswill JH, Parry JV, Clewley JP. Performance of the Amplicor human immunodeficiency virus type 1 PCR and analysis of specimens with false-negative results. J Clin Microbiol 1997; 35: 2846-53.
6. Fransen K, Van Kerckhoven I, Piot P, Van der Groen G. Evaluation and comparison of the Amplicor HIV-1 polymerase chain reaction kit with an "in-house" nested PCR. Clin Diagn Virol 1995; 4: 311-9.
7. Puthanakit T, Apichartpiyakul C, Sirisanthana V. An in-house HIV DNA PCR assay for early diagnosis of HIV infection in children in Thailand. J Med Assoc Thai 2003; 86: 758-65.
8. Van Laethem K, Beuselinck K, Van Dooren S, De Clercq E, Desmyter J, Vandamme AM. Diagnosis of human immunodeficiency virus infection by a polymerase chain reaction assay evaluated in patients harbouring strains of diverse geographical origin. J Virol Methods 1998; 70: 153-66.
9. Gómez Carrillo M, Avila M, Hierholzer J, et al. Mother-to-child HIV type 1 transmission in Argentina: BF recombinants have predominated in infected children since the mid-1980s. AIDS Res Hum Retroviruses 2002; 18: 477-83.
10. Carr JK, Avila M, Gómez Carrillo M, et al. Diverse BF recombinants have spread widely since the introduction of HIV-1 into South America. AIDS 2001; 15: F41-7.
11. Thomson MM, Delgado E, Herrero I, et al. Diversity of mosaic structures and common ancestry of human immunodeficiency virus type 1 BF intersubtype recombinant viruses from Argentina revealed by analysis of near full-length genome sequences. J Gen Virol 2002; 83: 107-19.
12. Thomson MM, Villahermosa ML, Vázquez-de-Praga E, et al. Widespread circulation of a BF intersubtype recombinant form among HIV-1-infected individuals in Buenos Aires, Argentina. AIDS 2000; 14: 897-9.
13. Mangano A, Pittis G, Galíndez C, Bologna R, Sen L. Reliability of laboratory markers of HIV-1 infection in Argentinean infants at risk of perinatal infection. AIDS Patient Care STDs 1998; 12: 691-6.
14. Kopka J, Batalla M, Mangano A, et al. Relevance of viral phenotype in the early AIDS outcome of pediatric HIV-1 primary infection. Pediatr Res 2002; 52: 475-80.
15. Batalla VM, Delucchi G, Cañizal AM, et al. Aplicación de la PCR anidada en el diagnóstico de infección por el HIV-1 en pediatría. Actualizaciones en SIDA 1997;5: 282-7.
16. Albert J and Fenyo EM. Simple, sensitive and specific detection of human immunodeficiency virus type 1 in clinical specimens by polymerase chain reaction with nested primers. J Clin Microbiol 1990; 28: 1560-4.
17. Heyndrickx L, Janssens W, Zekeng L, et al. Simplified strategy for detection of recombinant human immuno-deficiency virus type 1 group M isolates by gag/env heteroduplex mobility assay. J Virol 2000; 74: 363-70.
18. Delwart EL, Herring B, Rodrigo AG and Mullins JI. Genetic subtyping of human immunodeficiency virus using a heteroduplex mobility assay. PCR Methods Appl 1995; 4: S202-16.
19. Thomson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997; 25: 4876-82.
20. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 1999; 41: 95-8.
21. Kumar S, Tamura K and Jakobsen IB. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 2001; 17: 1244-5.
22. Aulicino PC, Kopka J, Mangano A, et al. Circulation of novel HIV-1 A, BC and F subtypes in Argentina. AIDS Res and Hum Retroviruses 2005; 21: 158-64.
23. Aulicino PC Kopka J, Rocco C, Mangano A, Sen L. Sequence analysis of a South-American HIV type 1 BC recombinant. AIDS Res Hum Retroviruses 2005; 21: 894-6.
24. McCutchan F. Understanding the genetic diversity of HIV-1. AIDS 2000; 14 (Suppl 3): S31- 44.
25. Rusell K, Carcamo C, Watts D, et al. Emerging genetic diversity of HIV-1 in South America. AIDS 2000; 14: 1785-91.
26. HIV sequence database. In: http://www.hiv.lanl.gov/content/hiv-db/mainpage.html [ Links ] [ Links ] [ Links ] [ Links ] [ Links ] [ Links ] [ Links ] [ Links ] [ Links ] [ Links ] [ Links ] [ Links ] [ Links ] [ Links ] [ Links ] [ Links ] [ Links ] [ Links ] [ Links ] [ Links ] [ Links ] [ Links ] [ Links ] [ Links ] [ Links ] [ Links ]
Recibido: 03-03-2006
Aceptado: 22-03-2006














