Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
Revista argentina de microbiología
versión impresa ISSN 0325-7541versión On-line ISSN 1851-7617
Rev. argent. microbiol. v.37 n.4 Ciudad Autónoma de Buenos Aires oct./dic. 2005
Evaluation of a nested-PCR assay for Streptococcus pneumoniae detection in pediatric patients with community-acquired pneumonia
C. Mayoral*, M. Noroña, M. R. Baroni, R. Giani, F. Zalazar
Sección Bacteriología, Práctica Profesional, Facultad de Bioquímica y Ciencias Biológicas, Universidad Nacional del Litoral, Av. Freyre 2150 (3000) Santa Fe, Argentina. *Correspondence: E-mail: cmayoral@fbcb.unl.edu.ar
ABSTRACT
The aim of the present work was to evaluate the usefulness of a simplified method for DNA extraction coupled to a nested-PCR protocol, based on the amplification of pneumolysin gene fragments for the diagnosis of pneumococcal pneumonia in pediatric patients with clinical and radiological evidence of bacterial infection. Bacterial DNA was extracted from sera by boiling and used without further purification in the PCR for the pneumolysin gene. None toxic reagents were used and the necessary steps to obtain the DNA were left at a minimum; furthermore, it overcomes the use of expensive commercial kits for DNA purification. The total procedure can be completed the same day of sampling and, most important, it avoids the use of sophisticated technology. Both in vitro analytical specificity and sensitivity (10 CFU/ml) of the assay were similar to those previously reported. When clinical samples were tested, the rate of positivity was shown to be 83.3% and 71% in pediatric patients with positive (group a) and negative blood cultures (group b), respectively. In group a, DNA detection was successful in samples from children without treatment or with less than 48 h of antibiotic therapy. None amplification was obtained from sera patients with viral infection or in samples from healthy controls. The application of the strategy described in this paper substantially seems to improve the diagnostic process in a determinate group: blood culture-negative children with pneumonia.
Key words: Streptococcus pneumoniae, pneumococcal pneumonia, nested-PCR
RESUMEN
Evaluación de una PCR anidada en pacientes pediátricos para diagnóstico de neumonía neumocócica adquirida en la comunidad. El objetivo del presente trabajo fue evaluar la utilidad de un método simplificado para extracción de ADN, acoplado a un protocolo de PCR anidada, basada en la amplificación de fragmentos del gen de la neumolisina para el diagnóstico de neumonía neumocócica en niños con evidencias clínicas y radiológicas de infección bacteriana. El ADN bacteriano fue extraído del suero por calentamiento y utilizado en la PCR para el gen de la neumolisina sin purificación posterior. Para la obtención de ADN no se utilizan reactivos tóxicos ni costosos "kits" comerciales. El procedimiento completo puede ser realizado en el día y lo que es más importante, evita el uso de tecnología sofisticada. La especificidad analítica in vitro y la sensibilidad (10 UFC/ml) del ensayo fueron similares a lo hallado en publicaciones anteriores. El porcentaje de muestras positivas fue del 83,3% y del 71% en los pacientes con hemocultivos positivos (grupo a) y negativos (grupo b), respectivamente. En el grupo a, sólo se obtuvieron resultados positivos mediante la PCR anidada en los pacientes no tratados o con menos de 48 hs de tratamiento antibiótico. No se obtuvieron señales de amplificación en los sueros de los pacientes con infecciones virales ni en las muestras del grupo control. La aplicación de la estrategia descripta incrementa la posibilidad diagnóstica de neumonía neumocócica en niños con hemocultivos negativos.
Palabras clave: Streptococcus pneumoniae, neumonía neumocócica, PCR anidada
INTRODUCTION
Streptococcus pneumoniae is the most important etiological agent of pneumonia worldwide (1, 10, 16). It is accepted that a definitive diagnosis requires the isolation of S. pneumoniae from blood or pleural fluid. However, in children with pneumonia, blood culture yields a sensitivity lower than 10 per cent due to the most of them presents no bacteraemia (9, 16) or due to a previous antibiotic therapy. Moreover, blood culture presents an additional drawback: they may take several days in order to give a positive result.
Tests such as immunological methods have been used to the diagnosis of this infectious disease in urine or serum samples, but they present both lower sensitivity and specificity (2, 3, 8, 11, 14, 16, 22).
Molecular methods such as Polymerase Chain Reaction (PCR) are used with increased frequency for the diagnosis of several infectious disease, including pneumo-coccal pneumonia, due to their high sensitivity, specificity and speed (4, 5, 7, 13, 20, 26). PCR has been used in selected groups of patients to detect S. pneumoniae in middle ear fluid from patients with acute otitis media (12, 25), in cerebrospinal fluid from patients with meningitis (6), and in serum or blood from patients with bacteraemia (8). In order to increase the sensitivity and specificity of the assays, several authors have applied other approaches such as nested-PCR in patients with bacteremic pneumococcal pneumonia with different results (21, 23, 24).
The aim of the present work was to evaluate the usefulness of a simplified method for DNA extraction coupled to a nested-PCR protocol based on the amplification of pneumolysin gene fragments for the diagnosis of pneumococcal pneumonia in pediatric patients with clinical and radiological evidence of bacterial infection.
MATERIAL AND METHODS
Subjects
Pediatrics patients (N= 105, age: 3 month to 5 years) admitted to the Hospital de Niños "Dr. Orlando Alassia" (Santa FE, Argentina) were included in this study. They were classified in the following groups:
a. Children with definitive diagnosis of pneumococcal pneumonia confirmed by blood-culture (N= 24).
b. Children with clinical and radiological evidence of bacterial pneumonia. The initial chest radiograph showed segmental, lobar or multilobar consolidation in all cases. All patients had negative blood-cultures (N= 75).
c. Children with respiratory disease of viral etiology (positive ELISA tests) (N=10).
d. Children without evidence of infection or active respiratory disease (N=20). They were admitted by different non infectious diseases at the same time as the patients with pneumonia, and they were matched for age (Control group).
Clinical samples
Serum samples were obtained from group (a) patients over the course of the illness when Streptococcus pneumoniae was isolated from blood culture: twenty patients were under antibiotic treatment for 24 to 48 h before sample collection. The remaining 4 patients were under antibiotic treatment during 72-120 h. Sera from patients in groups (b) and (c) were collected at the same time of the first sample for blood culture (without previous antibiotic therapy). Sera from control patients in group (d) were obtained at the time of admission at the hospital. All the serum samples were stored at -20 °C before DNA extraction.
DNA extraction from bacteria
Five to ten colonies were suspended in 100 µl of 0.1M HCl - Tris buffer pH 8.0 and incubated at 100 °C during 14 min. Then, a centrifugation at 10000 g for 1 min was carried out. The supernatant was collected and stored a 4°C until PCR assay.
DNA extraction from clinical samples
Serum (100 µl) was diluted with an equal volume of 0.1 M HCl-Tris buffer pH 8.0. The mixture was incubated at 100 °C for 14 min, centrifuged at 10000g during 5 min and the supernatant was stored at 4 °C until PCR assay. All serum samples were processed diluted 1:1 and 1:10 in 0.1M HCl-Tris buffer pH 8.0.
Polymerase chain reaction
Nested- PCR was performed using primers described by Salo et al (23) ; first-round (outer) PCR primers Ia (5-ATTTCTGTA-ACAGCTACCAACGA-3) and Ib (5-GAATTCCCTGTCTTTT-CAAGTC-3) amplify a 348-bp region. The second-round (inner) PCR primers IIa (5-CCCACTTCTTCTTGCGGTTGA-3) and IIb (5-TGAGCCGTTATTTTTTCATACTG-3) amplify a 208-bp region of the pneumolysin gene. In the first round PCR, 2,5 µl of DNA were added to the PCR mixture to give a final volume of 25 µl. Reaction mixture contained 1X Green Go-Taq PCR buffer (Promega Corp.), 20 pmol of each primer, 100 µM of dNTPs, 2.5 mM MgCl2and 1U of Taq DNA polymerase (Promega Corp.). Nested PCR was carried out using the same PCR mixture conditions with primers IIa and IIb, with 1µl of the first PCR transferred to a new tube containing 24 µl of PCR mixture.
Cycling parameters. DNA amplification (first and nested-PCR) was performed on a programmable thermal cycler (Progene, Techne, Cambridge, UK) with the following parameters: 94 °C for 10 min followed by 30 cycles of 94 °C for 30 sec, 55 °C for 30 sec, and 72 °C for 30 sec, with a last cycle at 72 °C for 7 min.
Analysis of PCR products. Ten microliters of PCR products were analyzed using electrophoresis on 2% agarose/1X TAE buffer gels in the presence of 0.5 µg/ml ethidium bromide at 510 V/cm during 35 min. Then, agarose gels were observed under UV illumination and photographed using orange filter.
Controls
Three different controls were included in PCR reactions. DNA extracted from S. pneumoniae ATCC 49619 was used as positive control. Serum samples from healthy adult volunteers were used as negative controls. Distilled water instead of DNA was used in the reagents controls.
Precautions to avoid DNA contamination
Multiple precautions were undertaken to ensure that no amplicon contamination of molecular reagents occurred. These precautions included: physical separation of the post-amplification manipulation procedures from the DNA extraction and master mix preparation, the use of barrier-filtered pipette tips, and extensive surface cleaning using hypochlorite and UV irradiation, in according to the Recommendations for Molecular Biology techniques (19).
Sensitivity and Specificity of the assay
In order to evaluate analytical sensitivity of the assays, serum samples from healthy donors with no recent history of pneumococcal disease were spiked with different amounts of a S. pneumoniae suspension to obtain final concentration of 10 to 106CFU/ml, verified by plate counting on 5% sheep blood agar medium. To determine the specificity of the assays, DNA extracted from a variety of non-pneumococcal bacteria (S. viridans, S. pyogenes, H. influenzae type b and K. pneumoniae) was used. Bacterial DNA extraction was carried out as above. In these experiments, an additional primer pair, which has broad specificity for the conserved eubacteria 16S rRNA sequences was used as amplification control (6). This primer pair amplifies a 370-bp fragment.
RESULTS
Specificity of the assay
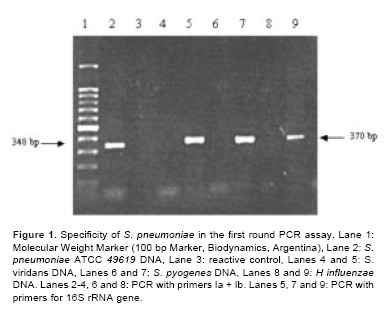
Figure 1 shows the results of specificity of PCR using DNA from S. pneumoniae and several non-pneumococcal bacteria. S. pneumoniae ATCC 49619 give the expected bands of 348 bp in the first round PCR. Conversely, no amplification products were obtained from the different non pneumococcal bacteria analysed. Furthermore, when 16S rRNA PCR primers were used, a proper fragment of 370 bp was amplified in all samples tested demonstrating the presence of DNA suitable to amplification. The expected band of 208 bp was obtained in the nested-PCR assay when S. pneumoniae ATCC was employed (data not shown).
Sensitivity of the assay
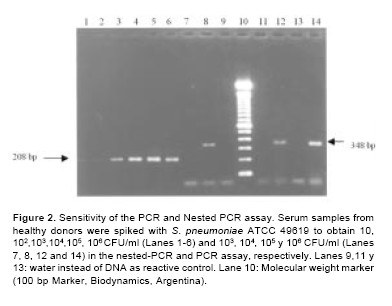
To determine the lowest limit of detection in these experiments, 10- fold serial dilutions of S. pneumoniae suspension in serum of healthy volunteers were tested by the protocol herein proposed. In PCR assay, 104CFU/ml give a detectable band in agarose gel electrophoresis. When nested-PCR was used to analyze the same dilutions, specific amplified products were obtained starting with 10 CFU/ml, which represents a 103- fold increase in the analytical sensitivity (Figure 2).
Detection of S. pneumoniae DNA in clinical specimens
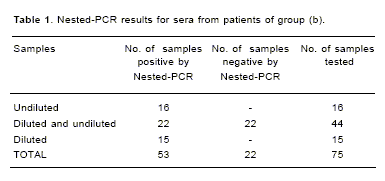
All serum samples were processed diluted 1:1 and 1:10 in 0.1M HCl-Tris buffer pH 8.0. Ten serum samples of group (a) were positive when first round PCR was applied. On the other hand, no amplification was obtained when we tested the samples from group (b). When the same samples were tested in the second round PCR (nested-PCR), 20/24 (83,3%) and 53/75 (71%) samples were positive in groups (a) and (b), respectively. None of the samples from the groups (c) and (d) were found to be positive by nested PCR. In some samples from group (b), the results were different if the sera were processed undiluted or diluted 1:10. Sixteen samples were positive undiluted, 15 were only amplificable after dilution, and 22 showed amplification undiluted or diluted 1:10 (Table 1). Five patients in group (b) had a previous antibiotic treatment. Two of them were under treatment at least 48 h before sampling and were nested-PCR negative. Remaining 3 patients (<48 h of antibiotic therapy) presented positive results.
DISCUSSION
In the present study, we evaluated a simplified diagnostic strategy for S. pneumoniae detection by DNA amplification in clinical samples from a pediatric patient population. It included a fast and safe method to extract DNA by boiling during 14 min. Unlike to other authors, none toxic reagents such as phenol or chloroform are used and the necessary steps to obtain the DNA are left at a minimum; furthermore, it overcomes the use of expensive commercial kits for DNA purification (4, 7, 13, 18, 24, 25, 26). In spite of its simplicity, when this method for DNA extraction is coupled to a nested PCR protocol based in that described by Salo et al. (22) with minor modifications, we achieve both analytical sensitivity and specificity similar to those reported in previous papers (21, 23). No amplification was observed in the presence of other related and non-related microorganism. Only ten CFU/ml were sufficient to give a detectable signal in agarose gel electrophoresis. After nested-PCR assay no consuming-time hybridization steps were used, as it is proposed in other papers (7, 25). When the clinical performance of the test was evaluated, we found a high rate of positive results (71%) in serum samples from pediatric patients whose first blood cultures were negative. Due to the favorable and rapid clinical evolution of patients in this group after empiric antibiotic treatment, a bacterial etiology of pneumonia could be assumed. None sample from patients of control groups included in the different assays gave positive results. As it has been pointed out by other authors, results in DNA amplification assays can be affected by the presence of PCR inhibitors (17, 24). Taking this into account, when a negative result was obtained using undiluted sample, the same was re-assayed starting from a 1:10 dilution. Using this approach, we have detected S. pneumoniae DNA in 15 samples after proper dilution. However, 22 samples from group (a) patients were negative by nested-PCR assay using undiluted or diluted samples. Several hypothesis could explain these results: i. incomplete removal of inhibitors by dilution, ii. antibiotic treatment before admission, iii. quick phagocytosis of bacteria by macrophages. Despite efforts to obtain samples prior to antibiotic administration in the hospital, several children (N=5, 6% in group (a) had received antibiotics before admission, samples of two of these patients gave negative result in the nested PCR (undiluted and diluted samples). In these patients the antibiotic treatment had been started at least 48 h before sampling. Similar results had been reported in previous studies (17). Total procedure can be completed the same day of sampling and the most important feature is that it avoids the use of sophisticated technology such as Real-Time PCR (15) and/or expensive DNA purification "kits". Finally, we believe that application of the strategy described in this study substantially improves the diagnostic process in blood culture-negative children with pneumonia, mostly in clinical laboratories from developing countries.
REFERENCES
1. Alonso de Velasco E, Verhoef J, Snippe H (1995) Streptococcus pneumoniae: Factors, pathogenesis and vaccines. Microbiol. Rev. 59: 591-603. [ Links ]
2. Congeni B, Igel H, Platt M (1984) Evaluation of a latex particle agglutination kit in pneumococcal disease. Pediatr. Infect. Dis. J. 3: 417-419. [ Links ]
3. Coovadia Y, Naidu K (1985) Evaluation of Bactigen latex agglutination and Phadecbact coagglutination for detection of bacterial antigens in cerebrospinal fluid. J.Clin. Pathol. 38: 561-564. [ Links ]
4. Dagan R, Shriker O, Hazan Y, Leibovitz E, Greenberg D, Schlaeffer F, et al (1998) Prospective study to determine clinical relevance of detection of pneumococcal DNA in sera of children by PCR. J. Clin. Microbiol. 36: 669-673. [ Links ]
5. Ehrlich G, Greenberg SJ (1994) PCR-based diagnostics for infectious disease. Blackwell Scientific Publications, Cambridge. [ Links ]
6. Greisen K, Loeffelholz M, Purohit A, Leong D (1994) PCR Primers and Probes for 16S r RNA gene of most species pathogenic bacteria, including bacteria found in Cerebrospinal Fluid. J. Clin. Microbiol. 32: 335-3351. [ Links ]
7. Hassan-King Y, Baldeh Y, Greenwood B (1994) Detection of Streptococcus pneumoniae DNA in blood culture by PCR. J. Clin. Microbiol . 32: 1721-1724. [ Links ]
8. Isaacman D, Zhang Y, Reynolds EA, Ehrlich GD (1998) Accuracy of a polimerase chain reaction-bassed assay for detection of pneumococcal bacteremia in children. Ped. 101: 813-816. [ Links ]
9. Johnston RB Jr (1991) Pathogenesis of pneumococcal pneumonia. Rev. Infect. Dis. 13 (Suppl. 6): S509-S517. [ Links ]
10. Klein JO (1981) The epidemiology of pneumoccocal disease in infants and children. Rev. Infect. Dis. 3: 246-253. [ Links ]
11. Lenthe-Eboa, Brighouse SG, Auckenthaler R (1987) Comparison of immunological methods for diagnosis of pneumococcal pneumonia in biological fluids. Eur. J. Clin. Microbiol. 6: 28-34. [ Links ]
12. Liederman EM, Post JC, Aul J (1998) Analysis of adult otitis media: polymerase chain reaction versus culture for bacteria and viruses. Ann. Otol. Rhinol. Laryngol. 107: 10-16. [ Links ]
13. Lorente ML, Falguera M, Nogués A, Ruiz Gónzalez A, Merino M, Rubio Caballero M (2000) Diagnosis of pneumococccal pneumonia by polymerase chain reaction (PCR) in whole blood: a prospective clinical study. Thorax. 55: 133-137. [ Links ]
14. Martin S, Hoganson D, Thomas E (1987) Detection of Streptococcus pneumoniae antigens in acute nonbacteremic pneumonia. J.Clin. Microbiol. 25: 248-250. [ Links ]
15. McAvin JC, Reilly PA, Roudabush RM, Barnes WJ, Salmen A, Jackson GW, et al (2001) Sensitive and specific method for rapid identification of Streptococcus pneumoniae using Real-Time Fluorescence PCR. J. Clin. Microbiol 39: 3446-3451. [ Links ]
16. Mufson MA (1995) Streptococcus pneumoniae. In Mandell G, Douglas, R, Bennett, J, Dolin, R, (Eds.), Principles and Practice of Infectious Diseases. 4th ed. Churchill Livingstone. New York, p. 1825-1828. [ Links ]
17. Murdoch DR, Anderson TP, Beynon KA, Chua A, Fleming AM, Laing RT, et al (2003) Evaluation of a PCR assay for detection of Streptococcus pneumoniae in respiratory and nonrespiratory samples from adults with community-acquired pneumonia. J.Clin. Microbiol. 41: 63-66. [ Links ]
18. Neumaier M, Braun A, Wagener C (1998) Fundamentals of quality assessment of molecular amplification methods in clinical diagnostics. Clin. Chem. 44: 12-26. [ Links ]
19. Peter JB (1991) The polymerase chain reaction: amplifying our options. Rev. Infect. Dis. 13: 166-171. [ Links ]
20. Rudolph KM, Parkinson A, Black CM, Mayer JLW (1993) Evaluation of polymerase chain reaction for diagnosis of pneumococcal pneumonia. J.Clin. Microbiol. 3l: 2661-2666. [ Links ]
21. Ruiz-González A, Nogués A, Falguera M (1997) Rapid detection of pneumococcal antigen in lung aspirates: comparison with culture and PCR technique. Respir Med. 91: 201-206. [ Links ]
22. Salo P, Ortqvist A, Leinonen M (1995) Diagnosis of bacteremic pneumococcal pneumonia by amplification of pneumolysin gene fragment in serum. J. Infect. Dis. 171: 479-482. [ Links ]
23. Toikka P, Nikkari S, Leinonen M, Mertsola J (1999) Pneumolysin PCR-based diagnosis of invasive pneumococcal infection in children. J. Clin. Microbiol 37: 633-637. [ Links ]
24. Virolainen A, Salo P, Jero J, Karma P, Eskola J, Leinonen M (1994) Comparison of PCR assay with bacterial culture for detecting Streptococcus pneumoniae in middle ear fluid of children with acute otitis media. J. Clin. Microbiol. 32: 2667-2670. [ Links ]
25. Zhang Y, Isaacman DJ, Wadowsky RM, Rydquist-White J, Post JC, Erlich GD (1995) Detection of Streptococcus pneumoniae in whole blood by PCR. J. Clin. Microbiol. 33: 596-601. [ Links ]
Aceptado: 14/11/05














