Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
Revista argentina de microbiología
versión impresa ISSN 0325-7541versión On-line ISSN 1851-7617
Rev. argent. microbiol. v.38 n.2 Ciudad Autónoma de Buenos Aires ene./abr. 2006
DNA fingerprinting by ERIC-PCR for comparing Listeria spp. strains isolated from different sources in San Luis, Argentina
A. Laciar*, L. Vaca, R. Lopresti, A. Vega, C. Mattana, O. N. P. de Centorbi
Área Microbiología, Facultad de Química, Bioquímica y Farmacia. Universidad Nacional de San Luis. Chacabuco y Pedernera (5700) San Luis, Argentina.
Correspondencia. E-mail: alaciar@unsl.edu.ar
ABSTRACT
In this study, a total of 24 Listeria spp. strains were analyzed. Twenty-two isolates were obtained in San Luis (Argentina) from human, animal, and food samples. Two types of strains, Listeria monocytogenes CLIP 22762 and Listeria innocua CLIP 74915, were included as reference strains. All isolates were biochemically identified and characterized by serotyping, phage typing, and amplification of the flaA gene by polymerase chain reaction (PCR). Repetitive intergenic consensus (ERIC) sequence-based PCR was used to generate DNA fingerprints. On the basis of ERIC-PCR fingerprints, Listeria spp. strains were divided into three major clusters matching origin of isolation. ERIC-PCR fingerprints of human and animal isolates were different from those of food isolates. In addition, groups I and II included ten L. monocytogenes strains, and only one Listeria seeligeri strain. Group III included nine L. innocua strains and four L. monocytogenes strains. Computer evaluation of ERIC-PCR fingerprints allowed discrimination between the tested serotypes 1/2b, 4b, 6a, and 6b within each major cluster. The index of discrimination calculated was 0.94. This study suggests that the ERIC-PCR technique provides an alternative method for the identification of Listeria species and the discrimination of strains within one species.
Key words: Listeria spp., ERIC-PCR, fingerprints DNA
RESUMEN
Caracterización molecular por ERIC-PCR de cepas de Listeria spp. aisladas de diversos orígenes en San Luis, Argentina. En este estudio se analizaron 24 cepas de Listeria spp. De ellas, 22 fueron obtenidas en San Luis (Argentina), a partir de muestras humanas, de animales y alimentos. Se incluyeron 2 cepas de referencia Listeria monocytogenes CLIP 22762 y Listeria innocua CLIP 74915. Todos los aislamientos fueron identificados bioquímicamente y caracterizados por serotipificación, fagotipificación y detección del gen flaA por reacción en cadena de la polimerasa (PCR). Se generaron perfiles de bandas de ADN mediante la amplificación de secuencias repetitivas de consenso intergénico de enterobacterias (ERIC-PCR). De acuerdo a los resultados obtenidos por ERIC-PCR, las cepas de Listeria spp. fueron divididas en 3 grupos según su origen. Los perfiles de los aislamientos humanos y animales fueron distintos de los correspondientes a alimentos. Por otra parte, dentro de los grupos I y II se incluyeron 10 cepas de L. monocytogenes y solamente una de Listeria seeligeri. Dentro del grupo III no sólo estuvieron incluidas las 9 cepas de L. innocua sino también 4 de L. monocytogenes. La evaluación de los perfiles de bandas obtenidos por ERIC-PCR permitió la discriminación entre los serotipos ensayados 1/2b, 4b, 6a y 6b dentro de cada grupo. El índice de discriminación calculado para ERIC-PCR fue de 0,94. Los resultados sugieren que la técnica de ERIC-PCR provee un método alternativo válido para la identificación de especies de Listeria y, asimismo, permite la diferenciación de cepas dentro de una misma especie.
Palabras clave: Listeria spp., ERIC-PCR, perfiles de ADN
INTRODUCTION
In the 1980s, several outbreaks of listeriosis were conclusively shown to be caused by the consumption of contaminated food (8) such as vegetables, milk, and meat products, in which these bacteria can multiply even at low temperatures. The presence of any Listeria species in food is usually an indication of poor hygiene (3). The coexistence of several species in the same food is not unusual, and the incidence of Listeria species other than Listeria monocytogenes is often higher than the incidence of L. monocytogenes itself. Because of the rising significance of the organism during the past decade, there has been increasing interest in developing fast, economical, and specific tests to detect L. monocytogenes in various types of food. However, one of the major problems has been the inability to distinguish L. monocytogenes from other non-pathogenic Listeria species, especially Listeria innocua, which may not only be present but also predominant in food samples (2). L. innocua, Listeria seeligeri, Listeria grayi, and Listeria welshimeri are considered to be generally avirulent. There is some evidence that suggests that strains of both L. seeligeri and L. innocua are occasionally pathogenic for man and animals, respectively (22, 24). L. seeligeri, previously described as experimentally nonpathogenic for mice, may in fact be a heterogeneous species regarding its pathogenicity, and includes strains that may cause life-threa-tening diseases in humans (23). Serotyping and phage typing have traditionally been used for the differentiation and characterization of Listeria spp. isolates (1, 25) since the vast majority of clinical isolates belongs to serotype 1/2a, 1/2b, or 4b, serotyping has limited value as an epidemiological tool (13). Phage typing has been useful in studies of some listeriosis outbreaks but it also has limited value, as not all strains are typeable (8, 21).
The use of several subtyping methods, including multilocus enzyme electrophoresis, total DNA restriction endonuclease analysis, ribotyping, pulsed-field gel electrophoresis (PFGE), and random amplified polymor-phic DNA analysis for sensitive subtyping of L. monocytogenes has been explored by various groups (10, 14, 18). The first studies of characterization of L. monocytogenes in food that exploited molecular methods date back to the 1990s. In the last years, only a few studies have considered the identification of the other members of the genus Listeria (7). The protocols developed are often able to detect only the genus Listeria, thus lacking the ability to identify different species of Listeria simultaneously (10).
Many molecular typing methods offer the advantage of high discriminating ability and typeability and do not require specialized reagents such as typing sera and bacteriophages. This characteristic has stimulated interest in the development of more rapid methods for the discrimination between Listeria spp. isolates such as PCR amplification of repetitive DNA sequences, which are dispersed in the genomes of bacteria in extragenic regions. Listeria spp. strains possess short repetitive extragenic palindromic (REP) elements and enterobacterial repetitive intergenic consensus (ERIC) sequences (13-15).
The purpose of this work was to determine whether DNA fingerprinting obtained by ERIC- PCR would allow discrimination between Listeria species and between different strains within one species.
MATERIALS AND METHODS
Bacterial isolates
A total of 24 strains of Listeria spp. was used in this study. The strains were isolated over the period 1994-2000 (Table 1). Two types of strains, L. monocytogenes (CLIP 22762) and L. innocua (CLIP 74915), were obtained from the Listeria Collection of the Pasteur Institute, Paris (France), and 22 isolates from San Luis (Argentina), isolated from humans (n=3), animals (n=3), and foods (n=16). All isolates were identified by Gram staining, catalase reaction, motility at 25 °C, and the Api Listeria kit (bioMérieux, Marcy I’Étoile, France) (5) at the time of isolation. They were further confirmed as genus Listeria by PCR detection of the flaA gene (6,11). The strains were serotyped and phage typed according to reference methods (16). All Listeria strains were maintained on Tryptone Soya Agar (Oxoid Ltd, Basingstake, Hants, England), and liquid cultures were grown in Brain Heart Infusion Broth (BHI) (Oxoid Ltd, Basingstake, Hants, England).
Serotyping
Serological identification was performed according to Seeliger et al. (25) based on the detection of 15 somatic antigens and five flagellar antigens that allow distinction of 13 serovars. This characterization is internationally used and recognized. The seric factors used were kindly provided by Dr. J. Rocourt (International Reference Center, Pasteur Institute, Paris).
Phage typing
Except for minor variations, the phage typing procedure closely followed previously published methods by Audurier et al. (1). Phages used were kindly provided by Dr. J. Rocourt.
DNA extraction
DNA was extracted from bacterial cells grown overnight at 37 °C in BHI by the method of Flamm et al. (9): that is, a buffer solution that contained 10 mM Tris-HCl, 1 mM EDTA (pH 8.0), 10% sodium dodecyl sulfate (Calbiochem), a proteinase K solution 20 mg/ml (Promega), and 0.25 mg/ml of lysozyme (Fluka, BioChemika) was added to the bacterial pellets. The mixture was incubated for 1 h at 37 °C, the enzymes were then inactivated by heating at 100 °C for 10 min. DNA was pelleted by centrifugation at 9,000 x g for 30 min and precipitated overnight at –20 °C with 2.5 volumes of absolute ethanol, and was allowed to dry in air. The pellets were suspended in 50 ml of sterile distilled water, and the DNA was quantified by measuring the optical density at 260 nm.
PCR amplification of the flaA and ERIC-PCR
PCR amplification of the flaA gene was performed by using the forward oligonucleotide primer (5’AGCTCTTAGCTCCATGAGTT3’) and the reverse primer (5’ACATTGTAGCTAAGGCGACT 3’) (6,11). For ERIC-PCR the forward primer (5’-ATGTAAGCTCCTGGGGA-TTCAC-3’) and the reverse primer (5’-AAGTAAGTGACTGGGG-TGAGCG) were used (13). Reactions were performed in a 50 μl volume in a Perkin Elmer thermocycler (Gene Amp PCR System 2,400). Reaction mixtures contained 1 μM of each primer, 50 a 100 ng of genomic DNA, 1.5 mM MgCl2; each deoxynucleoside triphosphate (dATP, dTTP, dCTP, dGTP; Promega, USA) at a concentration of 0.2 mM, and 1 U of Taq DNA polymerase (Promega, USA). For flaA amplification, 1 cycle at 94 °C for 2 min; 35 cycles at 94 °C for 1 min, 65 °C for 30 s, 72 °C for 30 s; and 1 cycle at 72 °C for 10 min were used. For ERIC-PCR amplifications were made with 1 cycle at 95ºC for 5 min, 30 cycles at 90 °C for 30 s, 50 °C for 30 s, 52 °C for 1 min, and 1 cycle at 72 °C for 8 min. All amplification products were resolved in agarose gels, stained with ethidium bromide, detected under a short-wavelength UV light source, and photographed with a Polaroid 667 camera. The 100bp DNA Ladder (Promega) was used as molecular size marker.
Computer-assisted analysis of the DNA fingerprints
All fingerprints were analyzed using the SYSTAT software version 5.0 (Systac Inc.), and the matrix of Ssm correlation coefficients was clustered by the unweighted paired group mean averages (UPGMA) method. Variations in intensity and shape of bands between isolates were not considered differences. The similarity between all fingerprints obtained by each method was expressed as a Dice coefficient correlation and was determined with the equation [2 nAB/(nA+nB)]x100, nAB being the number of matched fragments and nA+nB the total number of fragments in profiles A and B (19).
Discriminatory power
The discriminatory power of each fingerprinting method was estimated by the numerical index of discrimination (DI) calculated by the method of Hunter and Gaston (12), which is based on the probability that two unrelated strains sampled from the test population will be placed into different groups by the typing method.
The amplification assays were performed at least 3 times and were considered reproducible when identical genomic profiles were obtained in 3 separate experiments.
RESULTS
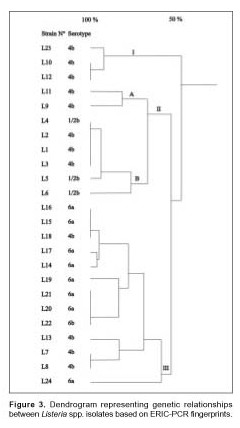
All Listeria spp. isolates were classified into four different serotypes, distributed as follows: 6a (n=8), ½ b (n=3), 4b (n=12), and 6b (n=1). The dates of isolation are listed in Table 1.
The amplification of a fragment of 450 bp corresponding to flaA gene confirmed the genus Listeria of the isolates (Figure 1).
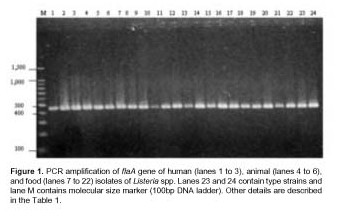
In the present study, all strains were examined by the ERIC-PCR technique. Visual comparison of banding patterns revealed 15 distinct ERIC profiles. Related strains (isolated from a mother and a newborn) produced identical ERIC profiles (Figure 2, lanes 1, 2 and 3). Each of the Listeria species yielded distinct fingerprints with multiple DNA fragments in sizes ranging between 150 and 1,570 bp (Figure 2). Common bands of 990 and 520 bp were observed in the twenty-four strains. Three common bands of 720, 460 and 150 bp were present in all the L. monocytogenes strains tested. All strains of L. innocua including type strain had two common bands of 1,570 and 1,340 bp. ERIC profiles of L. monocytogenes strains isolated from food were clearly distinct from ERIC profiles of human and animal L. monocytogenes strains (Figure 2).
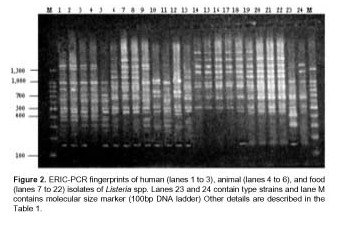
In contrast, one L. seeligeri animal isolate (L4) had a DNA banding pattern very similar to that of the three human isolates (L1, L2 and L3). In addition, one L. monocytogenes (L18) strain isolated from squid was included in the L. innocua group isolated from mussel. The estimated relationships between isolates on the basis of ERIC fingerprints are indicated on the dendrogram in Figure 3.
The percentage level of similarity clearly showed that the strains examined were divided into 3 distinct groups designated I, II and III. This first level was clustered at a similarity level of 50% and based on the origin of the Listeria strains (human, animal or food). Group I consisted of L. monocytogenes strains (serovar 4b, non typeable by phage typing) isolated from food in 1998. Group II subgroup A consisted of L. monocytogenes strains (serovar 4b, non typeable by phage typing) isolated from food. Subgroup B included L. monocytogenes strains isolated from humans and animals and one strain of L. seeligeri isolated from the intestinal content of Lagostomus maximus maximus in 1994 (17). Group III consisted of L. monocytogenes and L. innocua strains isolated from food over a three year period, 1997 to 2000. Within each of the three groups: I, II, and III, a further differentiation of ERIC profiles was established at 80% relative genetic similarity. This second level of clustering enabled us to obtain the same level of strain differentiation as serotyping, with the exception of L. seeligeri and L. monocytogenes serotype 1/2b. All L. innocua strains serotype 6a and one L. innocua strain serotype 6b were included in group III with the exception of four L. monocytogenes strains serotype 4b. Never-theless, there was a major differentiation of the L. monocytogenes strains identified as serovar 4b because seven different profiles were obtained by ERIC-PCR.
DISCUSSION
ERIC-PCR was used to characterize Listeria spp. strains isolated from humans, animals and foods. This method showed great possibilities for typing Listeria species.
Listeria spp. strains which are closely linked epidemiologically (human isolates of L. monocytogenes L1, L2 and L3) as well as L. seeligeri strain isolated from the intestinal content of L. maximus maximus showed very similar ERIC-PCR fingerprints by visual inspection. This is based on DNA homology values ranging between 1 and 28% of L. monocytogenes with other species of the genus, and on the coincidence of the peptidoglycan structure teichoic acids (17).
ERIC types correlated well with species and serotypes and generally showed greater differentiation within and between human, animal, and food isolates.
Among food isolates of L. monocytogenes and L. innocua, a great diversity of fingerprints was observed. ERIC profiles for food isolates were clearly different from the profiles obtained for human and animal isolates of L. monocytogenes (14).
The 24 Listeria spp. strains were divided into 3 groups, I, II, and III, clustered by a relative genetic similarity of 50%. This first level of clustering was grouped on the origin of the Listeria spp. strains (human, animal or food). The second level of clustering at 80% relative genetic similarity enabled us to obtain the same level of strain differentiation as serotyping, with the exception of L. seeligeri and L. monocytogenes serotype 1/2b which are more closely related to human isolates than to the other food isolates. In group III all strains were L. innocua serotype 6a and one serotype 6b with the exception of four L. monocytogenes strains serotype 4b. All these strains were isolated from food. All the strains tested in each genetic group were isolated the same year, except for L7, L8 and L13 belonging to group III, and L4 belonging to group II.
The strains were not typeable by phage typing in most cases, except L7 and L8 that showed the same phagovar and serovar, ERIC-PCR was unable to discriminate these strains and the profiles had a similarity of 100%.
The genomic profiles obtained with ERIC oligonucleotide allowed the molecular typing of each strain with 100% typeability and DI of 0.94. This index was lower than that observed by Boerlin et al using the RAPD technique with the combination of results obtained with two or three primers (4).
The division of the 24 strains of Listeria spp. by ERIC-PCR into a one group (II) containing human and animal isolates and another two groups (I and III) containing food isolates showed that no similarity between strains from humans or animals and strains from foods is present (Figure 3).
The ERIC-PCR technique applied on species of Listeria displayed a characteristic pattern that allowed to identify typical species bands. The bands obtained from L. monocytogenes were different from those obtained from L. innocua. The technique showed more discrimination than serotyping and phage typing. In fact, most strains were non-typeable by phage typing.
In conclusion, the results of this study based on ERIC-PCR showed that epidemiological investigations should include molecular typing of strains to clearly demonstrate whether a close genomic relationship exists among strains involved in outbreaks. Therefore, it can be claimed that the ERIC-PCR molecular technique may be useful as a Listeria spp. alternative typing system applied to elucidate listeriosis epidemiology.
REFERENCES
1. Audurier AMC. Phage typing of Listeria monocytogenes. Int J Food Microbiol 1989; 8: 251-7. [ Links ]
2. Carroll S, Carr L, Mallinson E, Lamichanne C, Rice B, Rollins D, et al. Development and evaluation of a 24-hour method for the detection and quantification of Listeria monocytogenes in meat products. J Food Prot 2000; 63: 347-53. [ Links ]
3. Cocolin L, Rantsiou K, Iacumin L, Cantoni C, Comi G. Direct identification in food samples of Listeria spp. and Listeria monocytogenes by molecular methods. Appl Environ Microbiol 2002; 68: 6273-82. [ Links ]
4. Boerlin P, Bannerman FIF, Rocourt J, Bille J. Typing of Listeria monocytogenes: a comparison of polymorphic DNA with 5 other methods. Microbiol Rev 1995; 46: 35-49. [ Links ]
5. Donnelly CW. Conventional methods to detect and isolate Listeria monocytogenes. In: Ryser ET, Marth EH, editors. Listeria, listeriosis, and food safety. New York, Marcel Dekker, Inc., 1999, p. 225-60. [ Links ]
6. Dons L, Rasmussen OF, Olsen JE. Cloning and characterization of a gene encoding flagellin of Listeria monocytogenes. Mol Microbiol 1992; 6: 2919-29. [ Links ]
7. Farber J, Addison C. RAPD typing for distinguishing species and strains in the genus Listeria. J Appl Bacteriol 1994; 77: 242-50. [ Links ]
8. Farber JM, Peterkin PI. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev 1991; 55: 476-511. [ Links ]
9. Flamm RK, Hinricks DJ, Thomashow MF. Introduction of pAMβ1 into Listeria monocytogenes by conjugation and homology between native Listeria monocytogenes plasmids. Infect Immun 1984; 44: 157-61. [ Links ]
10. Graves LM, Swaminathan B, Hunter SB. Subtyping Listeria monocytogenes, In: Ryser ET, Marth E H, editors. Listeria, listeriosis, and food safety. New York, Marcel Dekker, Inc., 1999, p. 279-97. [ Links ]
11. Gray DI, Kroll RG. Polymerase chain reaction amplification of the flaA gene for the rapid identification of Listeria spp. Lett Appl Microbiol 1995; 20: 65-8. [ Links ]
12. Hunter P, Gaston M. Numerical index of the discriminatory ability of typing systems: an application of Simson’s index of diversity. J Clin Microbiol 1988; 26: 2456-66. [ Links ]
13. Jersek B, Giltot P, Gubina M, Klun N, Mehle J, Tcherneva E, et al. Typing of Listeria monocytogenes strains by repetitive element sequence-based PCR. J Clin Microbiol 1999; 37: 103-9. [ Links ]
14. Jersek B, Tcherneva E, Rijpes N, Herman L. Repetitive element sequence based PCR for species and strain discrimination in genus Listeria. Lett Appl Microbiol 1996; 23: 55-60. [ Links ]
15. Jin LL, Wang QY, Hou X. ERIC-PCR on identification of Listeria species and strain. Yi Chuan 2003; 25: 195-7. [ Links ]
16. Laciar AL, Centorbi ON. Isolation and characterization of Listeria spp. isolated from seafood in San Luis (Argentina). Food Microbiol 2002; 19: 645-51. [ Links ]
17. Laciar A, Picca S, Centorbi ON. Aislamiento de Listeria seeligeri de ciego de vizcacha (Lagostomus maximus maximus). Rev Argent Microbiol 1994; 26: 183-8. [ Links ]
18. Nadon C, Woodward D, Young C, Rodgers F, Wiedmann M. Correlations between molecular subtyping and serotyping of Listeria monocytogenes. J Clin Microbiol 2001; 39: 2704-7. [ Links ]
19. Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA, 1979; 76: 5269-73. [ Links ]
20. Nogva HK, Rudi K, Naterstad K, Holck A, Lillehaug D. Application of 5´-nuclease PCR for quantitative detection of Listeria monocytogenes in pure cultures, water, skim milk, and unpasteurized whole milk. Appl Environ Microbiol 2000; 66: 4266-71. [ Links ]
21. Rocourt J, Audurier A, Courtieu L, Durst J, Ortel S, Schrettenbrunner P, et al. A multi-centre study on the phage typing of Listeria monocytogenes. Zbl Bakt Hyg 1985; 259: 489-97. [ Links ]
22. Rocourt J, Hof H, Schrettenbrunner A, Molinverni R, Bille J. Virulence moderee d’une souche humaine de Listeria seeligeri. In: Courtieu E, Espaze E, Reynaud A, editors. Listeriose, Listeria, Listeriosis, Proceedings of the 9th International Symposium on the Problems of Listeriosis, University of Nantes, Nantes University Press, 1986, p. 266-70. [ Links ]
23. Rocourt J, Hof H, Schrettenbrunner A, Molinverni R, Espaze E, Bille J. Meningité purulente aigue a Listeria seeligeri chez adulte inmunocompétent. Schweiz Med Wschr 1986; 116: 248-51. [ Links ]
24. Rocourt J, Seeliger H. Distribution des espéces du genre Listeria. Zbtralbl Bakteriol 1985; 259: 317-30. [ Links ]
25. Seeliger HPR, Höehne K. Serotyping of Listeria monocytogenes and related species. In: Bergan T , Norris J, editors. Methods in Microbiology. New York, Academic Press, 1979, p. 33-48. [ Links ]
Recibido: 13/6/05
Aceptado: 12/6/06














