Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
Revista argentina de microbiología
versión impresa ISSN 0325-7541versión On-line ISSN 1851-7617
Rev. argent. microbiol. v.41 n.1 Ciudad Autónoma de Buenos Aires ene./mar. 2009
Prevalence of mef and ermB genes in invasive pediatric erythromycin-resistant Streptococcus pneumoniae isolates from Argentina
A. Corso*, D. Faccone, C. Galiá, P. Gagetti, M. Rodríguez, J. Pace, M. Regueira,
THE "ARGENTINEAN SIREVA WORKING GROUP"
Servicio Antimicrobianos, Departamento Bacteriología, INEI - ANLIS "Dr. Carlos G. Malbrán", Av. Vélez Sarsfield 563, (1281) Ciudad Autónoma de Buenos Aires, Argentina.
*Correspondence. E-mail: acorso@anlis.gov.ar
This work was partially presented at the XVIth Lancefield International Symposium on Streptococci and Streptococcal disease. Palm Cove, Cairns, Australia, September 25-29, 2005.
Argentinean SIREVA Working Group: Cecilia Vescina, Hospital de Niños Sor Maria Ludovica, La Plata, Pcia. de Bs. As.; Martha Von Specht, Hospital Provincial de Pediatría, Posadas, Pcia. de Misiones, Diana Gómez and Victoria Monzani, Instituto Nacional de Epidemiología J. H. Jara/ Hospital Materno Infantil Don V. Tetamanti, Mar del Plata, Pcia. de Bs.As.; Clara Mayoral, Hospital de Niños Dr. O. Alassia, Santa Fe, Pcia. de Santa Fe; Andrea Nepote, Laboratorio Central de Santa Fe, Santa Fe, Pcia. de Santa Fe; Andrea Badano, Hospital de Niños Dr. V. J Vilela, Rosario, Pcia. de Santa Fe; Beatriz Garcia, Hospital H. Notti, Guaymallén, Pcia. de Mendoza; Claudia Hernández, Hospital de Pediatría SAMIC Dr. J P. Garrahan - CABA; Cristina Pérez, Hospital Provincial Castro Rendón, Pcia. de Neuquén; Sonia Flores, Hospital Enrique Vera Barros, La Rioja, La Rioja; Ana Littvik, Hospital Rawson, Córdoba, Pcia. de Córdoba; Maria José Rial, Hospital de Niños Dr. Pedro de Elizalde, CABA; Lydia Carvajal, Hospital de Niños Santísima Trinidad, Pcia. de Córdoba; Hortensia Cano, Hospital Regional de Río Gallegos, Río Gallegos, Pcia. de Santa Cruz; Julio Pace, Clínica Bazterrica, CABA; Adriana Procopio, Hospital de Niños Dr. Ricardo Gutiérrez, CABA, Argentina.
ABSTRACT
During the period 1993-2001, a total of 1,499 pneumococci isolates were recovered through the Argentinean surveillance of Streptococcus pneumoniae causing invasive disease in children under 6 years of age, 3.5% of which were erythromycin resistant. Among the 50 erythromycin-resistant strains available, 58% (n=29) harbored mefA/E genes (15 mefA, 30%; and 14 mefE, 28%), 34% (n=17) ermB, and 6% (n=3) both mefA/E plus ermB genes, while one isolate was negative for all the acquired genes studied. The England14-9 (42%), Poland6B-20 (20%) and Spain9v-3 (16%) clones were responsible for the emergence of pneumococcal macrolide resistance in pediatric population from Argentina.
Key words: Streptococcus pneumoniae; Macrolide; Argentina; Mef; erm.
RESUMEN
Prevalencia de los genes mef y ermB en aislamientos invasivos de Streptococcus pneumoniae resistentes a eritromicina recuperados de pacientes pediátricos en Argentina. En el marco del programa de vigilancia regional SIREVA, se analizaron 1499 aislamientos de Streptococcus pneumoniae causantes de enfermedad invasiva en menores de 6 años, recuperados entre 1993 y 2001. Se detectó un 3,5% de resistencia a eritromicina. De los 50 aislamientos resistentes a eritromicina que pudieron ser estudiados, el 58% (n=29) tenían los genes mefA/E (15 mefA, 30% y 14 mefE, 28%), el 34% (n=17) el gen ermB y el 6% (n=3) la combinación de genes mefA/E y ermB. Sólo un aislamiento fue negativo para todos los genes analizados. Los clones internacionales England14-9, Poland6B-20 y Spain9v-3 representaron el 78% del total de aislamientos resistentes (42, 20 y 16%, respectivamente) y se consideraron los responsables de la emergencia de la resistencia a macrólidos entre los neumococos que afectan a la población pediátrica de Argentina.
Palabras clave: Streptococcus pneumoniae; Macrólidos; Argentina; Mef; erm.
INTRODUCTION
Streptococcus pneumoniae is an important cause of morbidity and mortality in humans and one of the major causes of meningitis, community-acquired pneumonia and other respiratory tract infections (19). Over the last years, resistance to penicillin G in S. pneumoniae has increased in Latin America and worldwide (6, 11, 13). Consequently, macrolides are being used as initial empiric therapy for community-acquired respiratory tract infections (12). Unfortunately, resistance to macrolides in pneumococci has also evolved rapidly (3, 6, 9, 13, 15, 17) and a total of four different mechanisms of resistance have been described: i) antibiotic efflux, mediated by mefA/E genes, ii) targetsite modification by methylation of 23S rRNA due to erm genes, iii) mutations in domain II or V of 23S rRNA, and iv) mutations in riboproteins L4 or L22 (12, 17). The mefA/E genes have been associated with erythromycin resistance (14- and 15-membered macrolides) and display an M-phenotype (14, 17). The ermB gene is known to confer cross - resistance to macrolides, lincosamides and streptogramins B, yielding the MLSB-phenotype (12).
Since 1993, Argentina and other Latin American countries have participated in the Sistema Regional de Vacunas (SIREVA) Project, a regional epidemiological surveillance conducted by the Pan American Health Organization (11). The SIREVA Project focuses on the prevalence of capsular types and antimicrobial resistance patterns of S. pneumoniae causing invasive infections in children under 6 years of age. During the period 1993- 2001, a total of 1,499 invasive S. pneumoniae isolates were collected in Argentina through the SIREVA surveillance, 52 of which were erythromycin-resistant. Macrolide resistance emerged in Argentina in 1995 and increased from 0% in the period 1993-1994 to 1.4% in 1995-1997 and 6% in 1998-2001.
The aim of the present study was to determine both the prevalence of mefA/E and erm genes, and the clonal relationship among invasive erythromycin-resistant S. pneumoniae isolates.
MATERIALS AND METHODS
A total of 1,499 S. pneumoniae were submitted to the National Reference Laboratory from 39 hospitals, 17 provinces and Buenos Aires city. Fifty out of 52 erythromycin-resistant isolates from 14 hospitals (8 cities, 7 provinces) were available for further investigation. Thirty-one isolates (62%) were recovered from patients with pneumonia, twelve (24%) with meningitis, and seven (14%) with other infections. This subset of resistant isolates was recovered from blood (38%), pleural fluid (38%) and cerebrospinal fluid (24%). Eighty percent of isolates (n=40) were recovered from children ≤ 2 years old (0-6 months, 14 strains; 6-12 months, 12 strains; 1 year, 12 strains; and 2 years, 2 strains), the remaining 10 strains were from children ranging from >2 to <6 years old. Serotyping was determined by the capsular Quellung method using commercial antisera (Statens Seruminstitut, Copenhagen, Denmark) as recommended by the manufacturer. Minimal inhibitory concentrations (MICs) were determined by the agar dilution procedure using Mueller-Hinton agar (Difco, BD, USA) supplemented with 5% horse blood and incubated overnight at 35 ºC. MICs values to penicillin, cefotaxime, trimethoprim-sulfamethoxazole, tetracycline, chloramphenicol, ofloxacin, vancomycin, erythromycin, azithromycin, and clindamycin were interpreted according to CLSI guidelines (7). Isolates showing resistance to erythromycin but susceptibility to clindamycin expressed the Mphenotype, while isolates showing resistance to both erythromycin and clindamycin were grouped as having the MLSB-phenotype. Double disk diffusion assays using erythromycin (15 μg) and clindamycin (2 μg) disks (disk supplied by BBL, BD, USA) were performed in order to evaluate the inducible or constitutive expression of the MLSB-phenotype.
DNA templates were prepared by the boiling method and 5 μl of these extracts were used in each PCR assay. Reactions were performed under standard conditions in a final volume of 50 μl, using a Biometra thermal cycler (Whatman Biometra GmbH, Göttingen, Germany). Amplification of mefA/E, ermB, ermC, ermA and ermTR genes was performed with previously reported primers (24).
Discrimination between mefA and mefE alleles was performed by BamHI (New England Biolabs, Beverly, MA, USA) cleavage of the 348-bp mefA/E amplicon (17). Briefly, BamHI renders two fragments of 284 and 64 bp with the mefA allele, while mefE has no restriction sites.
PFGE was carried out using SmaI (New England Biolabs, Beverly, MA, USA) as previously described (8). DNA fragments were resolved in 1% agarose gel using a CHEF-DR III apparatus (Bio-Rad Laboratories, Hercules, CA, USA) by applying a switch time of 5 to 35 seconds during 23 h at 11.3 ºC (8). PFGE patterns were visually compared to 16 international clones described by McGee (16), kindly provided by Marguerite Lovgren (National Centre for Streptococcus, Edmonton, Alberta, Canada), and categorized using the Tenover criteria (25). MLST was performed at the Microbiology Laboratory, The Rockefeller University, NY, USA as previously described (27).
RESULTS AND DISCUSSION
Among the 50 erythromycin-resistant S. pneumoniae examined in this study, 25 (50%) and 8 (16%) isolates displayed non-susceptibility to penicillin and cefotaxime, respectively. Resistance to tetracycline and chloramphenicol was observed in 22 (44%) and 6 (12%) isolates, respectively. All isolates were susceptible to ofloxacin and vancomycin (Table 1). The MIC range for erythromycin and azithromycin was 4 - ≥ 512 μg/ml, while the range for clindamycin was 0.06-512 μg/ml. Isolates were grouped into those expressing the M-phenotype (n=29; 58%) or the MLSB-phenotype (n=21; 42%), and no induction was observed for the strains expressing the MLSB-phenotype (Table 1). Isolates displaying the MLSB-phenotype showed higher MICs to erythromycin, azithromycin and clindamycin than isolates with the M-phenotype, and were also associated with a high percentage of resistance to penicillin, tetracycline and chloramphenicol (Table 1).
Table 1. Susceptibility of S. pneumoniae isolates according to erythromycin-resistant phenotype.

All 29 M-phenotype isolates were positive for mefA/E genes (Table 2). Among the MLSB-phenotype isolates (n=21), 17 yielded PCR positive for ermB gene and 3 contained both mefA/E and ermB genes. One isolate displaying the MLSB-phenotype was repeatedly negative for mefA/E, ermB, ermC, ermA and ermTR acquired genes. Therefore, mutation in 23S rRNA or in riboproteins L4 or L22 might be a likely mechanism involved in this phenotype (12).
Table 2. Genotipic characterization and clonal relationship of erythromycin-resistant S. pneumoniae isolates.
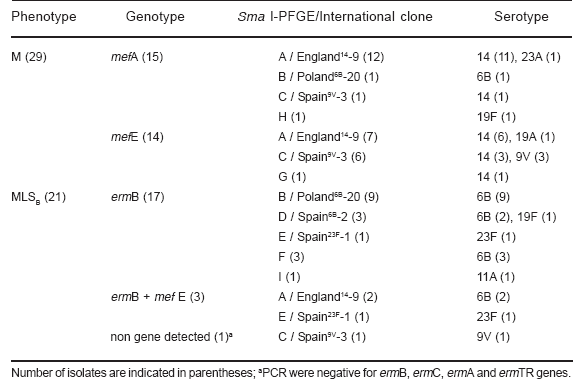
When we analyzed erythromycin resistance determinants in the complete set of 50 isolates, it was observed that 58% harbored the mefA/E genes, 34% the ermB and 6% both the mefA/E and the ermB genes. In North-American countries, the mef genes are more prevalent in S. pneumoniae (4, 12). In contrast, in Europe and South Africa, the ermB gene is the most frequent, while in Asian countries, the prevalence of genotypes is variable (12, 21, 23). Additionally, we determined by PCR-RFLP that 17 (53%) strains harbored the mefE allele, while the remaining 15 (47%), the mefA variant (Table 2). The mefA and mefE alleles are located in different genetic elements, Tn1207.1 and MEGA, respectively, showing different epidemiological behavior as described (14, 17). Tetracycline resistance in S. pneumoniae displaying the MLSB-phenotype has been associated with the presence of Tn1545, Tn1116, Tn3872 or Tn6002 transposons which encode tetM and ermB genes (5, 18). Although we did not look for presence of the tetM gene, 90% (n=18) of the S. pneumoniae isolates harboring the ermB gene were tetracycline- resistant, which suggests the presence of Tn1545 or Tn1545-like in Argentina (data not shown).
Serotype analysis (n; %) showed that the most frequent serotypes were: 14 (22; 44%) and 6B (17; 34%), while the remaining 11 S. pneumoniae expressed 6 different serotypes: 9V (4; 8%), 23F (2; 4%), 19F (2; 4%), 23A (1; 2%), 11F (1; 2%) and 19F (1; 2%) (Table 2). All 22 serotype 14 isolates expressed the M-phenotype, while 16 out of 17 serotype 6B strains harbored the ermB gene (MLSB-phenotype), two of which also contained the mefE gene (Table 2). We have previously reported that S. pneumoniae serotype 14 was the most frequent one causing invasive infections in pediatric patients (11, 22). Herein, we found that the emergence of erythromycin-resistant S. pneumoniae was mainly related to both serotypes 14 and 6B.
When analyzing the SmaI-PFGE pattern, 9 clonal types (A to I) were established (Figure 1, Table 2). Moreover, 88% of all isolates were related to 5 international clones (n; %): England14-9 (21; 42%), Poland6B-20 (10; 20%), Spain9v-3 (8; 16%), Spain6B-2 (3; 6%) and Spain23F-1 (2; 4%) (Table 2). Six strains, belonging to F-I patterns, were not related to any of the international clones used in this study as described by McGee et al. (Figure 1) (16). Eightyone percent (n=17) of the England14-9 clones were serotype 14, while all 10 Poland6B-20 isolates were serotype 6B (Table 2). Erythromycin-resistant Spain9v-3 isolates expressed serotypes 9V and 14 (Table 2). We and other authors have previously reported that the major penicillin- resistant S. pneumoniae clone in Argentina was the Spain9v-3 serotype 14 clone (1, 22). Additionally, MLST analysis of S. pneumoniae isolates from Brazil and Uruguay also described the association of the Spain9v-3 clone with serotype 14 (16, 20). Therefore, the presence of the Spain9v-3 clone expressing serotype 14 seems common in our region and suggests a capsular switching of 9V to 14. A genetic analysis between Spain9v-3 clones expressing 9V or 14 serotypes should be necessary to evaluate the origin and evolution of these clonal variants in Argentina. A representative isolate from the three major dominant clones was selected and typing was confirmed by MLST: England14-9 (ST 9), Poland6B-20 (ST 315) and Spain9v-3 (ST 156). In agreement with other authors, these three international clones were mainly implicated in the global dissemination of macrolide-resistance (10, 20, 26). Nineteen out of twenty-one England14-9 isolates harbored the mefA/E gene (12 mefA and 7 mefE), and 9 out of 10 Poland6B-20 strains carried the ermB gene in agreement with previous data (16, 20). Association was not found between serotypes or mef variants and geographical regions. The Spain9v-3 clone is usually susceptible to macrolides, but in this study and, in agreement with other authors, we found an association between this clone and the mef genes (2, 16).

Figure 1. SmaI-PFGE patterns of S. pneumoniae isolates. Marker: Lambda ladder PFG Marker (New England Biolabs). SmaIdigested DNA of S. pneumoniae R6 was also included as reference pattern. Panel A. Erythromycin-resistant S. pneumoniae clones determined in the present study, clones A to I. Clones A to E were related to some S. pneumoniae international clones. Panel B. S. pneumoniae international clones described by McGee et al. and in PMEN web site (16, 20).
In summary, 58% of the erythromycin-resistant isolates from the period 1993-2001 harbored the mefA/E gene, 34% the ermB gene, 6% contained the dual efflux (mefA/ E genes) plus methylase (ermB) mechanisms, and one isolate had neither gene. The emergence of erythromycin-resistant S. pneumoniae was principally due to the England14-9, Poland6B-20 and Spain9v-3 international clones.
Data from the Argentinean SIREVA surveillance of the period 2005-2006 showed that macrolide resistance reached 16.4% with a similar distribution of M- (57.9%) and MLSB-(42.1%) phenotypes (unpublished data). Moreover, almost 80% of these erythromycin-resistant S. pneumoniae belong to serotypes 14 (52%) or 6B (27%) (unpublished data), which suggests that nowadays both England14-9 and Poland6B-20 international clones are still the major clones involved in the spread of macrolide-resistance in Argentina. This is the first study both assessing the mechanisms of macrolide resistance in invasive pediatric S. pneumoniae isolates from Argentina, and the genetic relatedness between these organisms. Knowledge of the mechanisms and epidemiology of macrolide resistance in this pathogen is essential to predict the current and future use of these drugs as both first and second line antibiotics.
Acknowledgments: We thank José Luis Di Fabio and Jean-Marc Gabastou, Area of Technology and Health Service Delivery, Unit of Essential Medicines, Vaccines and Health Technology, PAHO Washington DC, USA. We also thank Marguerite Lovgren, National Centre for Streptococcus, Edmonton, Alberta, Canada, for providing the S. pneumoniae international clones, Prof. Alexander Thomasz and Herminia De Lencastre, Microbiology Laboratory, The Rockefeller University, NY, USA, for the MLST sequences. We thank Carla Duncan for her critical reading of the manuscript.
1. Albarracín Orio A, Cortés PR, Tregnaghi M, Piñas GE, Argentinean Network Pneumococcus Study Group, Echenique JR. A new serotype 14 variant of the pneumococcal Spain9v-3 international clone detected in the central region of Argentina. J Med Microbiol 2008; 57: 992-9. [ Links ]
2. Ardanuy C, Fenoll A, Berrón S, Calatayud L, Liñares J, the Spanish Pneumococcal Infection Study Network. Increase of the M phenotype among erythromycin-resistant Streptococcus pneumoniae isolates from Spain related to the serotype 14 variant of the Spain9v-3 clone. Antimicrob Agents Chemother 2006; 50: 3162-5. [ Links ]
3. Bonofiglio L, Ojeda MI, de Mier C, Vay C, Famiglietti A, Gutkind G, et al. Phenotypic and genotypic characterization of macrolide resistant Streptococcus pneumoniae recovered from adult patients with community-acquired pneumonia in an Argentinean teaching hospital. Int J Antimicrob Agents 2005; 25: 260-3. [ Links ]
4. Brown SD, Farrell DJ, Morrisey I. Prevalence and molecular analysis of macrolide and fluoroquinolone resistance among isolates of Streptococcus pneumoniae collected during the 2000-2001 PROTEKT US Study. J Clin Microbiol 2004; 42: 4980-7. [ Links ]
5. Calatayud L, Ardanuy C, Cercenado E, Fenoll A, Bouza E, Pallares R, et al. Serotypes, clones, and mechanisms of resistance of erythromycin-resistant Streptococcus pneumoniae isolates collected in Spain. Antimicrob Agents Chemother 2007; 51: 3240-6. [ Links ]
6. Castanheira M, Gales AC, Mendes RE, Jones RN, Sader HS. Antimicrobial susceptibility of Streptococcus pneumoniae in Latin America: results from five years of the SENTRY Antimicrobial Surveillance Program. Clin Microbiol Infect 2004; 10: 645-51. [ Links ]
7. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 16th informational supplement, 2006; M100-S16. Wayne, PA, USA. [ Links ]
8. Corso A, Severina EP, Petruk VF, Mauriz YR, Tomasz A. Molecular characterization of penicillin-resistant Streptococcus pneumoniae isolates causing respiratory disease in the United States. Microbial Drug Resist 1998; 4: 325-37. [ Links ]
9. Culasso C, Carvajal L, Paolucci R, Ceballos A, Paredes M. Streptococcus pneumoniae: evolution of antibiotic resistance in a pediatric hospital in Córdoba, Argentina. Rev Argent Microbiol 2001; 33: 149-54. [ Links ]
10. de la Pedrosa EGG, Morosini MI, van der Linden M, Ruiz-Garbajosa P, Galán JC, Baquero F, et al. Polyclonal population structure of Streptococcus pneumoniae isolates in Spain carrying mef and mef plus erm(B). Antimicrob Agents Chemother 2008; 52: 1964-9. [ Links ]
11. Di Fabio J, Castañeda E, Agudelo C, De La Hoz F, Hortal M, Camou T, et al. Evolution of Streptococcus pneumoniae serotypes and penicillin susceptibility in Latin America, Sireva.Vigía Group, 1993 to 1999. Pediatr Infect Dis J 2001; 20: 959-67. [ Links ]
12. Farrel DJ, Morrissey I, Bakker S, Felmingham D. Molecular characterization of macrolide resistance mecha-nisms among Streptococcus pneumoniae and Strepto-coccus pyogenes isolated from the PROTEKT 1999-2000 study. J Antimicrob Chemother 2002; 50 Suppl 1: S39-S47. [ Links ]
13. Felmingham D, Reinert RR, Hirakata Y, Rodloff A. Increasing prevalence of antimicrobial resistance isolates of Streptococcus pneumoniae from the PROTEKT surveillance study, and comparative in vitro activity of the ketolide, telitromycin. J Antimicrob Chemother 2002; 50 Suppl 1: S25-S37. [ Links ]
14. Klaassen CH, Mouton JW. Molecular detection of the macrolide efflux gene: to discriminate or not to discriminate between mef(A) and mef(E). Antimicrob Agents Chemother 2005; 49: 1271-8. [ Links ]
15. Klugman KP, Lonks JR. Hidden epidemic of macrolideresistant pneumococci. Emerg Infect Dis 2005; 11: 802-7. [ Links ]
16. McGee L, McDougal L, Zhou J, Spratt BG, Tenover FC, George R, et al. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the Pneumococcal Molecular Epidemiology Network. J Clin Microbiol 2001; 39: 2565-71. [ Links ]
17. Montanari MP, Mingoia M, Cochetti I, Varaldo PE. Phenotypes and genotypes of erythromycin-resistant pneumococci in Italy. J Clin Microbiol 2003; 41: 428-31. [ Links ]
18. Montanari MP, Cochetti I, Mingoia M, Varaldo PE. Phenotypic and molecular characterization of tetracyclineand erythromycin-resistant strains of Streptococcus pneumoniae. Antimicrob Agents Chemother 2003; 47: 2236-41. [ Links ]
19. Obaro S, Adegbola R. The pneumococcus: carriage, disease and conjugate vaccines. J Med Microbiol 2002; 51: 98-104. [ Links ]
20. Pneumococcal Molecular Epidemiology Network. PMEN. Clone collection. http://www.sph.emory.edu/PMEN. [ Links ]
21. Reinert RR, Reinert S, van der Linden M, Cil MY, Al-Lahham A, Appelbaum P. Antimicrobial susceptibility of Streptococcus pneumoniae in eight European countries from 2001 to 2003. Antimicrob Agents Chemother 2005; 49: 2903-13. [ Links ]
22. Rossi A, Corso A, Pace J, Regueira M, Tomasz A. Penicillinresistant Streptococcus pneumoniae in Argentina: frequent ocurrence of an internationally spread serotype 14 clone. Microbial Drug Resist 1998; 4: 225-31. [ Links ]
23. Song JH, Chang HH, Suh JY, Ko KS, Jung SI, Oh WS, et al. Macrolide resistance and genotypic characterization of Streptococcus pneumoniae in Asian countries: a study of the Asian Network for Surveillance of Resistant Pathogens (ANSORP). J Antimicrob Chemother 2004; 53: 457-63. [ Links ]
24. Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother 1996; 40: 2562-6. [ Links ]
25. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria of bacterial strain typing. J Clin Microbiol 1995; 33: 2233-9. [ Links ]
26. Wolter N, von Gottberg A, du Plessis M, de Gouveia L, Klugman KP, the Group for Enteric, Respiratory and Meningeal Disease Surveillance in South Africa (GERMSSA). Molecular basis and clonal nature of increasing pneumococcal macrolide resistance in South Africa, 2000-2005. Int J Antimicrob Agents 2008; 32: 62-7. [ Links ]
27. Zemlicková H, Crisóstomo MI, Brandileone MC, Camou T, Castañeda E, Corso A, et al. Serotypes and clonal types of penicillin-susceptible Streptococcus pneumoniae causing invasive disease in children in five Latin American countries. Microbial Drug Resist 2005; 11: 195-204. [ Links ]
Recibido: 07/10/08
Aceptado: 24/02/09.














