Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
Revista argentina de microbiología
On-line version ISSN 1851-7617
Rev. argent. microbiol. vol.42 no.3 Ciudad Autónoma de Buenos Aires July/Sept. 2010
MICROBIOLOGÍA CLÍNICA Y ENFERMEDADES INFECCIOSAS
Characterization of Suid herpesvirus 1 field isolates from Argentina
M. S. Serena1, 2 *, G. E. Metz1, 2, G. P. Martín Ocampos1, 2, S. E. Gambaro1, E. C. Mórtola3, M. G. Echeverría1, 2
1Cátedra de Virología,
3Cátedra de Inmunología, Facultad de Ciencias Veterinarias, Universidad Nacional de La Plata, 60 y 118, CC 296 (1900) La Plata, Argentina,
2CONICET (Consejo Nacional de Investigaciones Científicas y Técnicas).
*Correspondence. E-mail: solserena2000@fcv.unlp.edu.ar
ABSTRACT
The genomic characterization of Suid herpesvirus 1 (SHV-1) isolates from Argentina was accomplished by restriction pattern analysis using the BamHI, BstEII and XhoI enzymes. Type II genome has been described only once in Argentina. This study revealed considerable homogeneity of BamHI endonuclease sites in all the strains analyzed, according to the number and size of the fragments. No deletion of BamHI fragment #7 among the Argentinean isolates suggests that these strains are wild-type. In addition, the main antigenic domain of glycoprotein E of all the Argentinean strains, as well as the reference strains and sequences available in the GenBank, were characterized. The similarity percent oscillated between 99 and 100%.
Key words: Suid herpesvirus 1; RFLP; Glycoprotein E
RESUMEN
Caracterización de cepas de campo de Herpesvirus suino 1 aisladas en Argentina. Se caracterizaron cepas argentinas de herpesvirus suino tipo 1 (HVS-1) mediante corte con las enzimas de restricción BamHI, BstEII y XhoI. La presencia del tipo genómico II había sido reportada en la Argentina una sola vez y, hasta el momento, no se han informado nuevos casos. Este estudio reveló una homogeneidad de los sitios BamHI, acorde con el número y el tamaño de los fragmentos. La presencia del fragmento BamHI #7 en los aislamientos argentinos analizados sugiere que éstos pertenecen al tipo salvaje (wild-type). Además, se caracterizó el principal dominio inmunogénico de la glicoproteína E de todas las cepas argentinas y se lo comparó con los de las cepas de referencia y con las secuencias disponibles en la base de datos GenBank. Los porcentajes de similitud obtenidos oscilaron entre 99 y 100%.
Palabras claves: Herpesvirus suino 1; Patrones de restricción; Glicoproteína E
Aujeszky's disease (AD) or Pseudorabies (Pr) is one of the most significant infectious diseases in the pig industry. The causative agent, Suid herpesvirus 1 (SHV- 1), is indigenous in swine and is probably the cause of mass mortality among piglets. AD is manifested by various degrees of respiratory distress, nervous and genital disorders, and even mortality, according to the host age and the virulence of the viral strain involved. Also, after recovery, a latent infection is established. SHV-1, a member of the Alphaherpesvirinae subfamily, is a double-stranded DNA virus of approximately 90 megadaltons, composed of long and short unique sequences. The short sequence is bracketed by the internal and terminal repeat sequences (IR and TR respectively) and contains a group of four glycoprotein genes: gD, gE, gI and gG. This short sequence is located in the BamHI fragment #7 and is composed of 6.7 Kbp (10). Glycoprotein E, an envelope glycoprotein consisting of 577 amino acids (1.7 Kbp), is one of the SHV-1 virus virulence determinants, but is non-essential for in vitro replication. The main antigenic domain of gE has been identified between amino acids 52 and 84 (7). Field isolates of SHV-1 have been subdivided into genome types I, II and III, based on restriction pattern analysis using the BamHI enzyme (6). Thereafter, a subtyping has been carried out using other restriction enzymes, such as BstEII and XhoI: genome type I (Ia, Iia, Ib, In, Ip, Is and Iip), genome type II (IIa, IIp) and genome type III (IIIa and IIIp). RFLP of the SHV-1 genome has been used to analyze strains isolated in Argentina since 1994. At that moment, type I and II genome types of SHV-1 strains were known in Argentina, although the outbreak of genome type II has occurred only once so far (2). After that, we isolated five Argentinean strains from pigs with symptoms compatible with AD, coming from different regions of the country. In 1998, the Servicio Nacional de Sanidad Animal (SENASA) established a voluntary vaccination with an inactivated-gE-deleted vaccine. However, there is no information about the vaccine strains used in our country, because its importation was discontinued in 2001. The commercially available gE-deleted vaccine strains used worldwide include Bartha, NIA-3, NIA-4, Bucharest and Begonia strains (http://www.oie.int/esp/normes/mmanual/A_00041.htm). The latter is used in the Pr eradication program of Santa Catarina State in Brazil (13). The purpose of the presen study was to investigate the possible genetic variation among SHV-1 strains from Argentina by RFLP analysis in order to know if the strains are vaccine derivatives in comparison with non-deleted reference strains. In addition, we analyzed a partial sequence of the gE gene against the gE sequences available in the GenBank (Table 1). This is the first Argentinean study comparing sequences of SHV-1 strains.
Table 1. Viral isolates from GenBank

By using RFLP, we analyzed 13 SHV-1 strains, nine of which were isolated in Argentina from 1979 to 1998, and four have been previously characterized (RC/79, Mer, CL/7 and CL/15) (2). The non-deleted reference strains used were Ind-S (9), Alf (6) and S-66 (12), classified as type I, and YS-81, classified as type II (3). The CL/7, CL/15, TL/92, A/94, CL/96, CL/98 and CLP/98-10 strains were isolated in our laboratory, while the RC/79 and Mer strains were isolated in other Argentinean laboratories. All strains were propagated twice in Continuous Porcine Kidney (CPK) cell line cultures. The cells were grown in Eagle's medium (Gibco Invitrogen Corporation, USA) supplemented with 10% heat-inactivated fetal calf serum and antibiotics (penicillin-streptomycin). DNA was isolated from the cells infected with each viral strain at a multiplicity of infection between 1 and 5. CPK cells grown in 75 cm3 plastic flasks were inoculated with 1ml of supernatant of positive cell cultures, and after an extensive cytopathic effect was observed, pellets were washed and suspended in TEN buffer consisting of 100 mM Tris-HCl (pH7.5), 12.5 mM EDTA (pH 8.0), 150 mM NaCl and 1% SDS. Proteinase K at a final concentration of 0.2 mg/ml was then added and incubated at 50 °C for 4 h. The lysate was extracted once with TE buffer (10 mM Tris-HCl pH 8.0 and 1 mM EDTA pH 8.0), saturated phenol, and once with a mixture of phenol-chloroform-isoamylalcohol (25:24:1). Finally, total DNA was precipitated with two volumes of 99% ethanol, rinsed twice with 70% ethanol, dried and dissolved in 40 μl of sterilized distilled water. The quantity and quality of DNA was determined by measuring absorbance at an OD260/OD280 ratio in a spectrophotometer SmartSpec 3000 (Bio-Rad, USA). Approximately 1.5 μg of DNA solution from each viral strain was digested overnight with 2UI of BamHI at 37 °C, and then separated by electrophoresis in 0.7% (w/v) agarose gel (140 x 150 x 5 mm) in TAE buffer (40 mM Tris-acetate pH 7.8, 5 mM sodium acetate, and 1 mM EDTA) at 20 V for 16 h at room temperature, stained with ethidium bromide, visualized and photographed under a UV illuminator. After classifying all the strains as genome type I or II according to the BamHI pattern, the BstEII and XhoI enzymes were used to establish the subtypes respectively. The digestion reactions were carried out according to the conditions used for BamHI. Lambda DNA cleaved with HindIII and EcoRI + HindIII were used as size markers.
For Southern blotting (SB) analysis, electrophoretically separated DNA-fragments (1% w/v agarose) were transferred to nylon membranes (Hybond N+ membrane, Amersham, UK) in 20X SSC buffer (3 M sodium chloride, 300 mM sodium citrate, pH 7.0) and then prehybridized at 42 °C for 4 h in a solution containing 50% formamide, 6X SSC buffer (0.9 M sodium chloride and 0.09 M sodium citrate), 5X Denhardt's solution (0.1% bovine serum albumin -BSA-, 0.1% polyvinylpirrolidone, 0.1% Ficoll), 0.2% BSA, 1% sodium dodecyl sulfate -SDS-, 10 mM EDTA and 100 μg/ml denatured salmon sperm DNA. Probe hybridization proceeded overnight at 42 °C with the biotinylabeled BamHI fragment #7 originated from the Ind-S strain (Bio Prime DNA kit labeling system, BRL Life Technology) in hybridization buffer (40 mM Tris, 1 mM EDTA, 5X Denhardt's, 6X SSC, 0.1% BSA, 0.1% SDS). The chromogenic detection streptavidin-alkaline phosphatase method was used.
To amplify the main antigenic domain of gE (286 bp), we designed the following primers: ForgEpi (5'-CCC CGA GCC TCT CCG CCG A-3') and RevgEpi (5'-TCG CAG CCG CGG ACG GGC A-3'). PCR was performed in a DNA Thermal Cycler (Perkin-Elmer Cetus, Norwalk, CT, USA). A single PCR amplification was carried out according to the manufacturer's protocols using PCR Master Mix (Promega). Denaturation, annealing and extension consisted of 30 cycles at 94 °C for 30 s, 60 °C for 45 s and 72 °C for 1 min, respectively. PCR products were analyzed on 2.5% (w/v) agarose gels in TAE buffer, and stained with ethidium bromide. The products were visualized under UV light. The PCR products were purified according to the manufacturer's protocols using Wizard SV Gel and PCR Clean-Up System (Promega). PCR products were cloned into a pGEM-T Easy plasmid (Promega). After transformation of E. coli JM109 strain, the plasmid DNA was extracted and purified by the Wizard Plus Miniprep DNA Purification System (Promega). Two colonies of each strain were analyzed. Both chains of each cloned strain were sequenced using M13 standard primers by Mega BACETM 1000 (IGEVET, FCV-UNLP, La Plata, Argentina). Twelve sequences from different geographical origins were downloaded from the GenBank and were included in this analysis (Table 1). The sequence alignments and edition were carried out using the ClustalX version 1.92 (15) and BioEdit software version 5 (5). The percentage of similarity between sequences was calculated using the similarity index implemented in PAUP software (14).
According to the classification based on the presence of fragment 2 using BamHI (6), all Argentinean strains, except for the Mer strain, were classified into genome type I. Although the Mer strain (which is classified as type II) has only been isolated once so far, the five strains isolated after 1994 are still classified as type I. In the typical pattern II, an extra BamHI cleavage site, named 2a and 2b, appeared in fragment 2. Regarding type I, there were no alterations involving the gain or loss of BamHI cleavage sites in any of the strains analyzed. Although all Argentinean type I strains presented almost the same pattern, there were small differences and the main variation was in the 5+14', 10 and 12 fragments. In the Argentinean strains, as well as in the Alf strain, the 5+14' fragment was smaller than in the Ind-S and S-66 strains. Fragment 10 of strains RC/79 and CL/96 shifted its migration to the position of fragment 9; also fragment 10 presented different mobility from that of the others. In addition, fragment 12 was very variable in size in all the strains analyzed. In the subtyping analysis, BstEII was used to identify two subtypes: Is and Ip. Strains RC/79, CL/7, CL/15, TL/92, A/94 and CL/96 were classified as Is, because they showed an additional cleavage site in the BstEII fragment 2. In contrast, strains CL/98 and CLP/98-10 were classified as Ip. Taking into account BamHI and BstEII patterns, the Ind-S strain was classified into subtype Iia because its restriction pattern presented an additional cleavage site in fragment 5. Since the pattern of the S-66 strain was characterized by the lack of BamHI cleavage sites between fragments 14' and 5', S-66 was classified as subtype Iip. In addition, the Alf strain was classified as subtype Ib because its pattern showed an enlarged fragment 11 that co-migrated with fragment 10. Mer and YS-81 strains were verified as genome type II, subtype IIp. The restriction analysis using XhoI did not show different patterns between these two strains (Figure 1 and Table 2). When we analyzed the results obtained by SB analysis, we detected that fragment BamHI #7 of all strains, even type I or II, was in the same position and thus, not truncated.
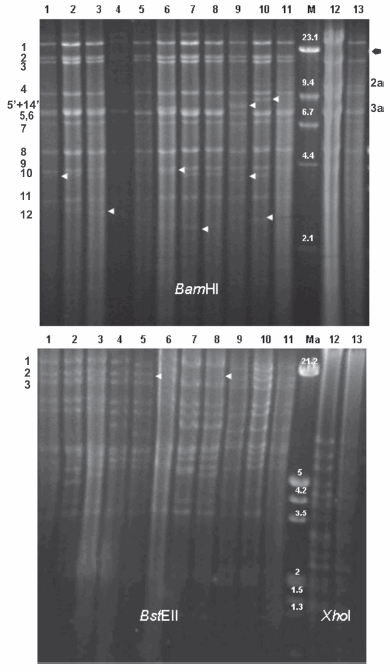
Figure 1. Restriction fragments obtained with the BamHI (upper), BstEII (lower left) and XhoI (lower right) enzymes. Lane 1: strain RC/79; Lane 2: strain CL/7; Lane 3: strain CL/15; Lane 4: strain TL/92; Lane 5: strain A/94; Lane 6: strain CL/96; Lane 7: strain CL/98; Lane 8: strain CLP/98-10; Lane 9: strain S-66; Lane 10: strain Ind-S; Lane 11: strain Alf; Lane 12: strain Mer; Lane 13: strain YS-81. Lane M, Lambda DNA cleaved with HindIII; Lane Ma, Lambda DNA cleaved with EcoRI + HindIII (size expressed in Kbp). Fragments are numbered following Herrmann et al. (13) for BamHI patterns type I (left numbers) and type II (right numbers). Fragments are numbered according to Christensen (1) for BstEII patterns (left numbers). Arrows indicated differences.
Table 2: Viral strains used in this work

Nd. Not determined
The percentage of similarity between gE nucleotide sequences of type I Argentinean strains and reference sequences Rice, Becker, S-66, Ind-S, Alf, Min-A and Ea was 100%. Besides, type I Argentinean strains showed 99.95% similarity with Fa, LA, Yangsan, Guangdong, RongA, GDSH and P-PrV strains. All Argentinean strains showed the lowest percentage of similarity (99.3%) with the PRV-SH strain. Likewise, type II Mer strain showed 100% similarity with its reference strain YS-81.
It is clear that all Argentinean SHV-1 strains are BamHI DNA pattern type I. The only strain isolated in Argentina previously classified as type II was isolated from animals imported from Holland (2) and was not disseminated in our country because all animals had been sacrificed. It has been reported that genomic type II is common in Holland (4) and that it is the predominant type in the pig population in Brazil (11, 13). The restriction patterns using BamHI were the same as those obtained by others for reference strains S-66, YS-81 and Ind-S (1, 8, 9), indicating that the passage in cell cultures made in our laboratory did not modify viral DNA. In agreement with the data obtained by others (4), the most variable BamHI fragments are 5 and 13, which are located within the repeat regions, and considerable variations can be seen in the sizes of BamHI fragments, 10, 12 and 8', which span the repeat-unique joint regions. In this study, the most variable fragments in Argentinean strains were 5+14', 10 and 12. No great variations were obtained between the other fragments. Those variations are known from viruses isolated from the same geographic area and are characterized by a different mobility. The Argentinean type I strains had a high degree of BamHI pattern identity to Alf and Ind-S strains, especially in fragment 5+14'. It is well known that the most noticeable change in the vaccine strain profile is the absence of BamHI fragment #7 (10). As reported by Schaeffer et al. (13), the vaccine strains NIA-4, Bartha, and Begonia could be grouped as type I, although they are significantly different in their DNA pattern because they have a deletion in this fragment. Even though this fragment is transcriptionally very active, it appears to be conserved and invariable in size among the high variety of field strains, as previously corroborated by others (1, 4). No deletion of BamHI fragment #7 was observed among the Argentinean strains, indicating that they are wild-type viruses. The differences in fragment position are relevant since they allow the follow-up of viral spread within the porcine population and the possible identification of vaccine strains. In reference to subtyping, six strains (RC/79, CL/7, CL/15, TL/92, A/94, CL/96) were classified as subtype Is and two (CL/98, CLP/98-10) as subtype Ip, whereas the Mer strain was classified as IIp. This study revealed the absence of type In and type III in Argentina. When we analyzed the sequence of the fragment encoding the main antigenic domain of the gE protein, we found that the differences between the genome type I and type II strains are not significant. Due to the high similarity detected between analyzed sequences, future studies aiming to carry out a phylogenetic analysis of SHV-1 strains from Argentina, will benefit from choosing other more variable regions. In summary, the Argentinean viruses were wild type and were classified as type I, suggesting that no major variation has occurred in SHV-1 spreading in Argentina since the first outbreak was noticed. No new isolates belonging to genome type II have been reported since 1981.
Acknowledgements: The assistance of Ms. Mondragón, Ms. A. Conde and Mr. C. Leguizamón is highly acknowledged. This study was supported by a FONCyT grant (PICT 2003 01-13451).
1. Christensen L. The population biology of suid herpesvirus 1. APMIS Supp 1995; 48: 1-48. [ Links ]
2. Echeverría M, Norimine J, Galosi C, Oliva G, Etcheverrigaray M, Nosetto E. The genotype of Aujeszky's disease viruses isolated in Argentina. J Vet Med Sci 1994; 56: 985-7. [ Links ]
3. Fukusho A, Shimizu M, Kjubo M, Namba K, Shimizu Y, Konno S, et al. The first outbreak of Aujeszky's disease in swine in Japan. Bull Natl Inst And Health 1981; 85: 5-11. [ Links ]
4. Gielkens A, Van Oirschot J, Berns A. Genome differences among field isolates and vaccine strains of pseudorabies virus. J Gen Virol 1985; 66: 69-82. [ Links ]
5. Hall T. BioEdit: a user friendly biological sequence alignment editor and analysis program for Window 95/98/NT. Nucleic Acids Sym Ser 1999; 41: 95-8. [ Links ]
6. Herrmann S, Heppner B, Ludwig H. Pseudorabies virus from clinical outbreaks and latent infections grouped into four major genome types. In: Wittmann G, Gaskell R, Rziha H, editors. Latent herpesvirus infections in veterinary medicine, current topics in veterinary medicine and animal science. Boston, USA. Martinus Nujhoff Pub, 1984, p. 387-401. [ Links ]
7. Jacobs L, Meloen R, Gielkens A, Van Oirschot J. Epitope analysis of glycoprotein I of pseudorabies virus. J Gen Virol 1990; 71: 881-7. [ Links ]
8. Nishimori T, Imada T, Sakurai M, Kitabayashi T, Kawamura H, Nakajima H. Restriction endonuclease analysis of Aujeszky's disease viruses isolated in Japan. Jpn J Vet Sci 1987; 49: 365-7. [ Links ]
9. Paul P, Mengeling W, Pirtle E. Differentiation of pseudorabies (Aujeszky's disease) virus strains by restriction endonuclease analysis. Arch Virol 1982; 73: 193-8. [ Links ]
10. Petrovskis E, Timmins J, Gierman T, Post L. Deletions in vaccine strains of pseudorabies virus and their effect on synthesis of glycoprotein gp63. J Virol 1986; 60: 1166-9. [ Links ]
11. Piatti R, Akimi Ikuno A, Sequetin Cunha E, D'Ambros R, Gregori F, Martins Soares R, et al. Characterization of Aujeszky's disease virus isolates from South and Southeast Brazil by RFLP analysis. Braz J Microbiol 2001; 32: 144-6. [ Links ]
12. Rockborn G, Hyllseth B. The second outbreak of pseudorabies in Sweden. Nord Vet Med 1968; 20: 193-6. [ Links ]
13. Schaefer R, Ciacci-Zanella J, Mores N, Pan K, Feltrim Dambros R, Fracasso Schiochet M, et al. Characterization of Aujeszky's disease virus isolated from South Brazil in the last twenty years by restriction enzyme analysis. Braz J Microbiol 2006; 37: 390-4. [ Links ]
14. Swofford D. PAUP*. Phylogenetic analysis using parsimony (*and others methods). Version 4. 1998; Editorial Sinauer Associates, Sunderland. [ Links ]
15. Thompson J, Gibson T, Pleuniak F, Jeanmougin F, Higgins D. Multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997; 25: 4876-82. [ Links ]
Recibido: 17/02/10
Aceptado: 19/05/10














