Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
Revista argentina de microbiología
Print version ISSN 0325-7541
Rev. argent. microbiol. vol.42 no.4 Ciudad Autónoma de Buenos Aires Oct./Dec. 2010
INDUSTRIAL AND ENVIRONMENTAL MICROBIOLOGY
Optimization of killer assays for yeast selection protocols
C. A. Lopes, M. P. Sangorrín*
Laboratorio de Microbiología y Biotecnología, Instituto Multidisciplinario de Investigación y Desarrollo de la Patagonia Norte (IDEPA), Consejo Nacional de Investigaciones Científicas y Técnicas-Universidad Nacional del Comahue, Buenos Aires 1400, (8300) Neuquén, Neuquén, Argentina.
*Correspondence. E-mail: sangorrinmarcela@conicet.gov.ar
ABSTRACT
A new optimized semiquantitative yeast killer assay is reported for the first time. The killer activity of 36 yeast isolates belonging to three species, namely, Metschnikowia pulcherrima, Wickerhamomyces anomala and Torulaspora delbrueckii, was tested with a view to potentially using these yeasts as biocontrol agents against the wine spoilage species Pichia guilliermondii and Pichia membranifaciens. The effectiveness of the classical streak-based (qualitative method) and the new semiquantitative techniques was compared. The percentage of yeasts showing killer activity was found to be higher by the semiquantitative technique (60%) than by the qualitative method (45%). In all cases, the addition of 1% NaCl into the medium allowed a better observation of the killer phenomenon. Important differences were observed in the killer capacity of different isolates belonging to a same killer species. The broadest spectrum of action was detected in isolates of W. anomala NPCC 1023 and 1025, and M. pulcherrima NPCC 1009 and 1013. We also brought experimental evidence supporting the importance of the adequate selection of the sensitive isolate to be used in killer evaluation. The new semiquantitative method proposed in this work enables to visualize the relationship between the number of yeasts tested and the growth of the inhibition halo (specific productivity). Hence, this experimental approach could become an interesting tool to be taken into account for killer yeast selection protocols.
Keywords: Killer yeast; Killer capacity; Biocontrol; Semi-quantitative methods; Spoilage yeast
RESUMEN
Optimización de la actividad killer para protocolos de selección de levaduras. En este trabajo se presenta un nuevo ensayo semicuantitativo que optimiza la detección de actividad killer en levaduras. Se evaluó la actividad killer de 36 cepas pertenecientes a las especies Metschnikowia pulcherrima, Wickerhamomyces anomala y Torulaspora delbrueckii, en vista del potencial uso de estas levaduras como agentes de biocontrol frente a las especies contaminantes de vinos Pichia guilliermondii y Pichia membranifaciens. Se comparó la efectividad de la técnica clásica basada en estrías (método cualitativo) con la del nuevo método semicuantitativo. El porcentaje de levaduras que mostraron actividad killer fue más alto cuando se utilizó el método semicuantitativo (60%) que con el método cualitativo (45%). En todos los casos, el agregado de 1% de NaCl en el medio permitió una mejor observación del fenómeno killer. Se observaron importantes diferencias en la capacidad killer de diferentes cepas dentro de la misma especie. Se detectaron dos cepas de W. anomala (NPCC 1023 y 1025) y dos cepas de M. pulcherrima (NPCC 1009 y 1013) con un amplio espectro de acción, ya que fueron capaces de inhibir el desarrollo de las tres levaduras sensibles evaluadas. La evidencia experimental demuestra la importancia de una adecuada selección de la cepa sensible al evaluar la actividad killer. El nuevo método semicuantitativo propuesto en este trabajo permite visualizar la relación entre el número de levaduras sembradas y el halo de inhibición del crecimiento (productividad específica). En conclusión, este método resulta una herramienta interesante para ser tenida en cuenta en los protocolos de selección de levaduras killer.
Palabras clave: Levaduras killer; Capacidad killer; Biocontrol; Método semicuantitativo; Levaduras contaminantes
INTRODUTION
Killer yeasts can produce toxic proteins or glycoproteins (killer toxins) that can cause death in sensitive (killer-sensitive) yeast isolates. The killer phenotype appears to be widely distributed within many yeast genera isolated from different sources (10, 36, 37). The interest in the killer property for industrial and medical applications has recently increased (11, 15, 22); however, the ecological role of killer yeasts has been somewhat overlooked and the conditions governing their behavior in particular habitats are mostly unknown (8).
The killer phenomenon is important in some industrial alcoholic fermentation processes using non-sterile substrates, even in those conducted by selected yeast starters in which wild yeast isolates may interact (17, 26, 28, 36). Killer isolates influence the fermentation process (by provoking changes in the population balance) and cause, in some cases, the unsuccessful implantation of a selected yeast isolate and the subsequent alterations in the final product (40, 43).
On the other hand, food spoilage caused by microorganisms is a serious problem for the food industry (20, 38). The exploration of killer yeasts as producers of mycocinsactive against these undesired microorganisms seems interesting. Hence, the use of selected killer yeasts as a biocontrol method may be related to the improvement of the food industry by reducing the requirement for chemical preservatives (5, 9, 33). In previous studies carried out in our laboratory, the majority of yeasts isolated from spontaneously fermenting grape must have evidenced the killer character (18, 29, 30, 33). The high variability in the killer phenomenon detected within different producer species provides an exceptional source of antagonist yeasts to be used in biocontrol. In particular, killer yeasts belonging to Metschnikowia pulcherrima, Wickerhamomyces anomala (ex Pichia anomala) and Torulaspora delbrueckii species have shown the broadest killer spectrum against wine spoilage yeasts (19, 33).
As a general rule, most studies on killer behaviour use the seeded-agar-plate technique based on the screening procedure described by Makower and Bevan (23) due to its simplicity and low cost (1, 2, 31, 41). In this traditional qualitative (QLM) experimental approach, yeasts to be tested for sensitivity are seeded as a lawn and the yeasts to be assayed for killer activity are streaked over the surface of the solidified YEPD-MB agar medium. Killer activity is then evidenced by the presence of a growth inhibition halo of the target yeast around the killer yeast streak.
Some slight modifications in the composition of the killer medium were reported, including the addition of glycerol or NaCl as well as variations in pH and temperature (7). However, these modifications were not adopted in general routine killer assays. The use of NaCl in the killer medium was particularly proposed for halotolerant yeasts isolated from olive brines, soy sauce, pickled radish, etc. (11, 16, 39).
The aim of the present work was to develop an alternative rapid and effective semiquantitative killer assay for biocontrol yeast selection. The effect of the addition of different NaCl concentrations as well as the use of serial dilutions of each killer yeast on the detection and comparison of different killer yeasts against spoilage yeasts was evaluated.
MATERIALS AND METHODS
Yeast isolates
Thirty six killer yeasts: 14 M. pulcherrima, 13 W. anomala and 9 T. delbrueckii isolates, from the North Patagonian Culture Collection (NPCC, Neuquén, Argentina) were used (Table 1). All killer yeasts used in this work were selected from screening surveys on yeasts isolated from wine related environments (19, 30, 33). In addition, we analyzed the killer activity of five reference strains including the type isolates of the three species previously mentioned (Table 1).
Table 1. Autochthons and reference killer yeast strains.
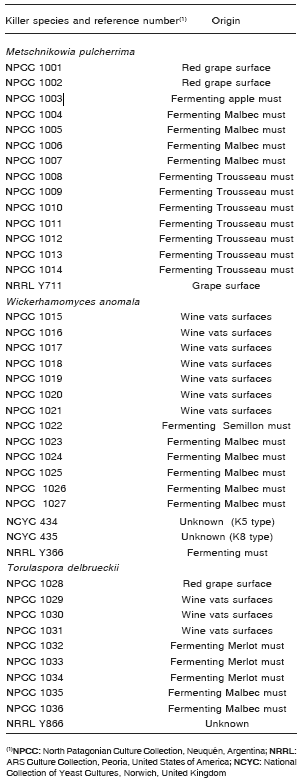
Three target strains were used: Pichia guilliermondii NPCC 1037 and Pichia membranifaciens NPCC 1108 (both species classified as potential wine spoilage yeasts) and Candida glabrata NCYC 388 (sensitive reference isolate).
Detection of killer activity
Each killer yeast isolate was tested for its ability to kill the three selected target isolates by a classical qualitative (QLM) and by a new semiquantitative (QTM) killer detection method.
In both cases, the seeded-agar-plate technique was used. In this technique, each target yeast isolate is suspended in sterile water (106 cells/ml) and 1 ml of this suspension is seeded as a lawn onto 10 ml of molten YEPD-MB agar plates (g/l: yeast extract 1, malt extract 3, glucose 20, peptone 2, agar 20, methylene blue 0.003, buffered at pH 4.6 with 0.5 M phosphate-citrate). Different NaCl concentrations (0, 1 or 3% w/v) were added to the killer medium in order to evaluate its effect in the improvement of killer activity detection. Young cultures (48 h in YEPD agar at 26 °C) of both sensitive and killer yeasts were used for killer assays. All experiences were carried out in triplicate.
In QLM, the seeded plates were streaked with thick smears of killer cultures and incubated at 18 ± 2 °C for 48-72 h. The lawn target yeast isolate was designated as sensitive when a clear zone of growth inhibition was observed surrounding the killer cultures streaks (23).
In QTM, 48 h cultures of all isolates to be tested for killer activity were resuspended in sterile water (1010 cells/ml) and five serial decimal dilutions were carried out in sterile water. Five microliters of all six consecutive dilutions were spotted on seeded plates and incubated at 18 ± 2 °C for 48-72 h. The diameter of the inhibition halo around the spot was used as a measurement of the killer activity. Halo areas for each sensitive/killer isolates combination were measured on digitalized images (Nikon Image Software (NIS) ver. 3.00), as previously reported (3).
Killer yeasts were evaluated according to two different concepts: (i) effectiveness, defined as the percentage of isolates showing killer activity against a particular sensitive isolate over the total number of yeasts tested, and (ii) specific productivity, defined as a relationship between the production of toxins (size of the inhibition halo) and the number of killer cells assayed (aliquots of known concentration).
The evaluation of dose-response results (killer cell counts vs. inhibition halo in mm) was carried out by lineal regression (Sigma Plot 2000).
RESULTS AND DISCUSSION
According to previous studies carried out in our laboratory, as well as in other laboratories wordwide, M. pulcherrima, W. anomala and T. delbrueckii killer yeasts species showed a significant inhibitory effect on the growth of different spoilage yeasts (6, 27, 32, 33).
In order to evaluate the effect of NaCl addition on killer behaviour, the three killer species previously described were evaluated against two wine spoilage yeast strains belonging to species P. guilliermondii (strain NPCC 1037) and P. membranifaciens (strain NPCC 1108) as well as against the sensitive reference strain Candida glabrata NCYC 388. The results using the QLM evidence an increase in halo size in the presence of 1 and 3% (w/v) NaCl with regard to the control (Table 2). Moreover, in some cases killer activity was evidenced only when NaCl was added to the medium (Table 2) (e.g. killer activity of M. pulcherrima against C. glabrata), indicating that the addition of a NaCl percentage to the killer medium can be an interesting factor to be taken into account for killer yeast screening.
Table 2. Killer activity (mm) at different NaCl concentrations of M. pulcherrima, W. anomala and T. delbrueckii type strains against target strains by QLM.

Qualitative methodological procedure (QLM) offers some economy of labor when a large number of yeasts are handled. However, it has been wrongly utilized in many studies for the definition of weak or strong phenotypes according to the extent of the halo produced by the streak (14, 26, 40, 42). We consider that this is an incorrect extrapolation of the methodology for the evident variation in the quantity of cells in the killer yeast streak. Based on this issue, we propose a new semiquantitative method (QTM) to evaluate killer activity which may provide a new and improved tool for selection of yeast isolates to be used as potential biocontrol agents. Killer yeasts in known concentrations were assayed using aliquots of six serial decimal dilutions from 1010 to 105 cells/ml on YEPD-MB agar plates containing the same sensitive isolates and the same NaCl concentrations previously used in the QLM (Table 3). As a general rule, a gradual increase of the growth inhibition halo was observed proportionally with the increasing amount of killer cells (Table 3 and Figure 1). Again, the addition of NaCl allowed a clearer visualization of the halos; however a delay in the growth of target yeasts in the medium supplemented with 3% NaCl was also exhibited. As shown in Table 3, the highest correlation values between dose (killer cell counts) and response (inhibition halos in mm) were obtained with the addition of 1% NaCl. According to these results, the optimum amount of 1% (w/v) of NaCl was selected to be added to the YEPD-MB medium used in all subsequent killer assays.
Table 3. Growth inhibition halo (mm) of killer strains evaluated at different NaCl concentrations (%) by QTM.

Figure 1. Growth inhibition halo of killer strains evaluated at different concentrations 1010-105 cells/ml; against P. guilliermondii NPCC 1037 a; P. membranifaciens NPCC 1108 b; and C. glabrata NCYC 388 c; by QTM.
In order to evaluate the effectiveness of this new QTM method in discriminating the best killer yeast to be potentially used in biocontrol protocols, a total of 41 killer yeasts (Table 1) were evaluated against the regional spoilage yeasts previously mentioned. In all the cases, the percentage of yeasts showing killer activity was found to be higher by the semiquantitative method (60% of the total number of crosses considering 1010 cells/ml) than by the qualitative method (45% of the total number of crosses) (Table 4). According to our results, it seems to be evident that the relationship between killer and sensitive cells has a great influence on the detection of the killer activity (Figure 1 and Table 4). Moreover, the percentage of yeasts showing killer activity when the QLM method was used resulted similar to the percentage detected at 108-109 cells/ml in the QTM method (Table 4).
Table 4. Effectiveness of QLM and QTM methods

Agar well diffusion bioassay using cell free culture supernatants or concentration disk bioassays have been used by several authors to detect and quantify killer activity (3, 27, 10). In these kinds of methodologies, thorough studies are needed regarding the relative stability of purified killer toxins after long term storage including the establishment of the actual time course of their possible degradation or loss of effectiveness (4). The semiquantitative method is a cost and time saving protocol which makes possible to analyze the cell/toxin ratio without toxin preparations. Our results evidenced that QTM is a useful method to approach the specific productivity of each isolate since it represents a relationship between the number of yeasts tested and the growth of the inhibition halo.
A different spectrum of action and specific productivity were evidenced when the activity of the three killer species against the three target yeasts was evaluated by using the QTM method (Figures 2 and 3). When the killer species were tested against the spoilage isolate P. guilliermondii NPCC 1037, killer activity percentages ranging from 100% for the species W. anomala and 30% for T. delbrueckii were observed (Table 4). Thirty seven percent of W. anomala isolates evidenced a maximum inhibition halo diameter over 5 mm when the highest concentration of killer yeasts (1010 cells/ml) was evaluated (Figure 2). In particular the W. anomala isolates NPCC 1018, 1022 and NRRL Y866 showed killer activity even at the lowest yeast concentrations (105 cells/ml), evidencing high specific productivity.
Figure 2. Inhibition halo size of killer isolates evaluated at different concentrations against P. guilliermondii NPCC 1037. The Arabic numbers indicate the particular isolate number according to Table 1.

Figure 3. Inhibition halo size of killer isolates evaluated at different concentrations against Pichia membranifaciens NPCC 1038. The Arabic numbers indicate the particular isolate number according to Table 1.
Low effectiveness of QTM at maximum concentration evaluated (1010 cell/ml) of killer yeasts species W. anomala (13%) and T. delbrueckii (0%) against spoilage species P. membranifaciens was observed (Figure 3 and Table 4). Seventy seven percent of M. pulcherrima killer isolates tested proved to be effective against P. membranifaciens, evidencing a higher effectiveness of this species against P. membranifaciens than against P. guilliermondii spoilage yeasts. Contrarily, W. anomala killer yeasts showed a higher effectiveness against P. guilliermondii than against P. membranifaciens (Figures 2 and 3).
On the other hand, 95% of the panel of killer yeasts was effective against the sensitive reference strain C. glabrata NCYC 388. Only isolates M. pulcherrima NPCC 1008 and W. anomala NPCC 1017 did not show killer activity against the sensitive reference strain C. glabrata; but these isolates proved to be killer against the spoilage yeasts (Figures 2 and 3). Moreover, in 60% of the cases tested in this work the inhibition halo produced by the killer yeasts was bigger against C. glabrata than against the spoilage yeasts. Taking into account that specific productivity and diffusion capacity are the same when the same killer isolate and the same medium are used, the width of growth inhibition halo observed among the tested killer isolate indicates that sensitive yeasts do not interact in the same way with the same killer toxin. In agreement with the current literature (6, 34), which seems to indicate a mechanism of possible toxin binding until saturation of free receptors, the rationale of this behavior could be found in the different number of free receptors available on sensitive cell walls for interaction. As a matter of fact, several authors suggested that the type and number of free active sites differ among different sensitive isolates (3, 12). Because of its high sensitivity, C. glabrata NCYC 388 has been used as reference sensitivity strain for killer yeast screening since the early studies of Young (44). We confirmed these observations and we are in agreement with the use of this isolate for general killer yeast screenings. However, as each killer yeast has a specific spectrum of action, we strongly recommend the use of those yeasts to be biocontrolled as the target isolate in killer assays.
As mentioned above, important differences were observed in different isolates belonging to a same killer species. These differences, which were not detected by the QLM method, are of relevance for the selection of the best suitable biocontrol agent. Among M. pulcherrima killer yeasts, isolates NPCC 1009 and 1013 presented the widest killer spectrum since they are the only ones to inhibit the three yeasts tested. Although pulcherriminic acid synthesized by this species has demonstrated antimicrobial effect (21, 25), no inhibitory activity of this compound was evidenced in the present study. In many cases a brown halo, produced as a consequence of the diffusion of pulcherriminic acid, was observed surrounding the killer colonies with no inhibitory effect against the target yeasts (Figure 1 c).
Among W. anomala killer yeasts, isolates NPCC 1023 and 1025 killed the three yeasts tested. An interesting observation is the fact that W. anomala isolates from fermentation musts (isolates NPCC 1022 to 1027) showed bigger diameters in the inhibition halos than those produced by the isolates isolated from grapes (isolates NPCC 1015 to 1021). This behaviour was particularly evident when P. guilliermondii (Figure 2) and C. glabrata (Figure 4) were used as target yeasts. This difference in killer phenotype regarding the origin of the isolates is in agreement with previous studies evidencing that P. guilliermondii yeasts isolated from grapes were found to be more sensitive to killer toxins than yeasts of the same species isolated from fermenting musts (19). This result seems to indicate that in an environment enriched with killer yeasts (fermenting must) the surviving isolates are mostly resistant to killer toxins.
Figure 4. Inhibition halo size of killer isolates evaluated at different concentrations against C. glabrata NCYC 388. The Arabic numbers indicate the particular isolate number according to Table 1.
Finally, the three T. delbrueckii isolates (NPCC 1030, 1034 and NRRL Y866) which were effective against P. guilliermondii also exhibited the highest specific productivity against C. glabrata. All these differences at intra-specific level detected in the three killer species tested may be due to the production of different amount of toxins or different toxins by each particular isolate (24).
The QTM method proposed in this work enables to visualize the relationship between the number of yeasts tested and the growth of the inhibition halo (specific productivity) in most of the cases. We conclude that, as a primary screening, the QLM method may be used to determine killer activity in a high number of yeasts although its effectiveness may vary depending on the target yeast used, the addition of salts or the number of killer yeasts (Table 3). After this first screening, the QTM method may be used to evaluate the yeast isolates evidencing killer activity in order to select the one showing the best performance.
1. Aguiar C, Lucas C. Yeasts Killer/Sensitivity Phenotypes and Halotolerance. Food Technol Biotechnol 2000; 38: 39-46. [ Links ]
2. Boone C, Sdicu AM, Wagner J, Degre R, Sánchez C, Bussey H. Integration of the yeast K1 killer toxin gene into the genome of marked wine yeasts and its effect on vinification. A J Enol and Vitic 1990; 41: 37-42. [ Links ]
3. Buzzini P, Martini A. Differential growth inhibition as a tool to increase the discriminating power of killer toxin sensitivity in fingerprinting of yeasts. FEMS Microbiol Lett 2000; 193: 31-6. [ Links ]
4. Buzzini P, Turchetti B, Vaughan-Martini AE. The use of Killer sensitivity patterns for biotyping yeast isolates, the state of the art, potentialities and limitations. FEMS Microbiol Lett 2007; 7: 749-60. [ Links ]
5. Comitini F, Ingeniis J, Pepe L, Mannazzu L, Ciani M. Pichia anomala and Kluyveromyces wickerhamii killer toxins as new tools against Dekkera/ Brettanomyces spoilage yeasts. FEMS Microbiol Lett 2004; 238: 235-40. [ Links ]
6. De Ingeniis J, Raffaelli N, Ciani M, Mannazzu I. Pichia anomala DBVPG 3003 secretes a ubiquitin-like protein that has antimicrobial activity. Appl Environ Microbiol 2009; 75: 1129-34. [ Links ]
7. Golubev W I. Mycocins killer toxins. In: Kurtzmann CP, Fell JW,. editors. The Yeasts, A Taxonomic study. Amsterdam, Elseiver Sci. Publ, 1998 p. 55-62. [ Links ]
8. Golubev WI. Antagonistic interactions among yeasts. In: Gábor P, Rosa C, editors. Biodiversity and Ecophysiology of Yeasts. Berlin, Heiderlberg, Springer, 2006, p. 187-219. [ Links ]
9. Goretti M, Turchetti B, Buratta M, Branda E, Corazzi L, Vaughan-Martini A, et al. In vitro antimycotic activity of a Williopsis saturnus killer protein against food spoilage yeasts. Int J Food Microbiol 2009; 131: 178-82. [ Links ]
10. Gulbiniene G, Kondratiene L, Jokantaite T, Serviene E, Melvydas V, Petkuniene G. Occurrence of killer yeast isolates in fruit and berrywine yeast populations. Food Technol Biotechnol 2004; 42: 159-63. [ Links ]
11. Hernández A, Martín A, Córdoba M G, Benito MJ, Aranda E, Pérez-Nevado F. Determination of killer activity in yeasts isolated from the elaboration of seasoned green table olives. Int J Food Microbiol 2008; 121: 178-88. [ Links ]
12. Hodgson VJ, Button D, Walker GM. Anti-candida activity of a novel killer toxin from yeast Williopsis mrakii. Microbiology 1995; 141: 2003-12. [ Links ]
13. Izgu F, Altinbay D. Killer toxins of certain yeast isolates have potential growth inhibitory activity on Gram-positive pathogenic bacteria. Microbios 1997; 89: 15-22. [ Links ]
14. Kapsopoulou K, Barouti E, Makrioniti A, Kostaki K. Occurrence of Saccharomyces cerevisiae killer yeasts in wine-producing areas of Greece. World J Microbiol Biotechnol 2008; 24: 1967-71. [ Links ]
15. Liu SQ, Marlene Tsao M. Inhibition of spoilage yeasts in cheese by killer yeast Williopsis saturnus var saturnus. Inter J Food Microbiol 2009;131: 280-2. [ Links ]
16. LLorente P, Marquina D, Santos A, Peinado JM, Spencer-Martins I. Effect of salt on the killer phenotype of yeasts from olive brines. Appl Environ Microbiol 1997; 63: 1165-67. [ Links ]
17. Lopes CA, Rodríguez ME, Sangorrín MP, Querol A, Caballero AC. Patagonian wines, the selection of an indigenous yeast starter. J Indus Microbiol Biotechnol 2007; 348: 539-46. [ Links ]
18. Lopes CA, Rodríguez ME, Sangorrín MP, Querol A, Caballero AC. Patagonian wines, implantation of an indigenous isolate of Saccharomyces cerevisiae in fermentations conducted in traditional and modern cellars. J Indus Microbiol Biotechnol 2007; 34: 139-49. [ Links ]
19. Lopes CA, Sáez JS, Sangorrín MP. Differential response of Pichia guilliermondii spoilage isolates to biological and physical-chemical factors prevailing in Patagonian wine fermentations. Can J Microbiol 2009; 55: 801-9. [ Links ]
20. Loureiro V, Malfeito-Ferreira M. Spoilage yeasts in the wine industry. Int J Food Microbiol 2003; 86: 23-50. [ Links ]
21. MacDonald J C. Biosynthesis of Pulcherriminic. Acid. Biochem J 1965; 96: 533-8. [ Links ]
22. Magliani W, Conti S, Travassos L R, Polonelli L. From yeast killer toxins to antibiobodies and beyond. FEMS Microbiol Lett 2008; 288: 1-8. [ Links ]
23. Makower M, Bevan EA. The inheritance of a killer character in yeast Saccharomyces cerevisiae. In: Geerts N editor. Genetics Todays, Proceedings of the 11 th International Congress of Genetics, MacMillan, The Hague, 1963, p. 202-3. [ Links ]
24. Marquina D, Santos A, Peinado JM. Biology of killer yeasts. Int Microbiol 2002; 5: 65-71. [ Links ]
25. Miller MW, Phaff HJ. Metschnikowia Kamienski. In: Kurtzman CP, Fell JP, editors. The Yeasts. Amsterdam, Elsevier 1998, p. 256-67. [ Links ]
26. Musmanno RA, Di Maggio T, Coratza G. Studies on strong and weak killer phenotypes of wine yeasts, production, activity of toxin in must, and this effect in mixed culture fermentation. J Indus Microbiol Biotechnol 1999; 87: 932-8. [ Links ]
27. Palpacelli V, Ciani V, Rossini G. Activity of different killer yeasts on isolates of yeast species undesirable in the food industry. FEMS Microbiol Lett 1991; 168: 75-8. [ Links ]
28. Pérez F, Ramírez M, Regodón JA. Influence of killer isolates of Saccharomyces cerevisiae on wine fermentation. Antonie van Leeuwenhoeck 2001; 79: 393-9 [ Links ]
29. Rodríguez ME, Lopes CA, van Broock MR, Vallés S, Ramón D, Caballero, AC. Screening and typing of Patagonian wine yeasts for glycosidase activity. J Appl Microbiol 2004; 96: 84-95. [ Links ]
30. Sangorrín MP, Zajonskovsky I, Lopes CA, Rodríguez ME, van Broock MR, Caballero AC. Killer behaviour in wild wine yeasts associated with Merlot and Malbec type musts spontaneously fermented from Northwestern Patagonia Argentina. J Basic Microbiol 2001; 41: 105-13. [ Links ]
31. Sangorrín MP, Zajonskovsky I, van Broock MR, Caballero AC. The use of killer biotyping in an old patagonian winery yeast ecological survey. World J Microbiol Biotechnol 2002; 18: 115-20. [ Links ]
32. Sangorrín MP, Lopes CA, Giraudo MR, Caballero AC. Diversity and killer behaviour of indigenous yeasts isolated from the fermentation vat surfaces in four Patagonian wineries. Int J Food Microbiol 2007; 119: 351-7. [ Links ]
33. Sangorrín MP, Lopes CA, Jofré V, Querol A, Caballero AC. Spoilage yeasts associated with Patagonian cellars, characterization and potential biocontrol based on killer interactions. World J Microbiol Biotechnol 2008; 24: 945-53. [ Links ]
34. Santos A, Marquina D, Barroso J, Peinado JM. 1-6-b-D glucan as the cell wall binding site for Debaryomyces hansenii killer toxin. Lett Appl Microbiol 2002; 34: 95-9. [ Links ]
35. Santos A, San Mauro M, Bravo E, Marquina D. PMKT2, a new killer toxin from Pichia membranifaciens, and its promising biotechnological properties for control of the spoilage yeast Brettanomyces bruxellensis. Microbiology 2009; 155: 624-34. [ Links ]
36. Schaffrath R, Meinhardt F. Kluyveromyces lactis zymocin and other plasmid-encoded yeast killer toxins. In: Schmitt MJ, Schaffrath R, editors. Microbial Protein Toxins, Berlin Heidelberg, Springer-Verlag, 2005, p. 133-55. [ Links ]
37. Schmitt MJ, Breinig F. The viral killer system in yeast, from molecular biology to application. FEMS Microbiol Rev 2002; 26: 257-76. [ Links ]
38. Stratford M. Food and beverage spoilage yeast. In: Querol, A, Feet G, editors. Yeasts in food and beverages II. Germany, Berlin Heidelberg, Springer-Verlag, 2006, p. 335-80. [ Links ]
39. Suzuki C, Yamada K, Okada N, Nikkuni S. Isolation and characterization of halotolerant killer yeasts from fermented foods. Agric Biol Chem 1989; 53: 2593-7. [ Links ]
40. Vagnoli P, Musmanno RA, Cresti S, Di Maggio T, Coratza G. Occurrence of killer yeasts in spontaneous wine fermentations from the Tuscany region of Italy. Appl Environ Microbiol 1993; 59: 4037-43. [ Links ]
41. Van Vuuren HJJ, Jacobs CJ. Killer yeasts in the wine industry, a review. Am J Enol Vitic 1992; 43: 119-28. [ Links ]
42. Walker G, McLeod A, Hodgson V. Interactions between killer yeast and pathogenic fungi. FEMS Microbiol Lett 1995; 127: 213-22. [ Links ]
43. Yap NA, de Barros Lopes M, Langridge P, Henschke PA. The incidence of killer activity of non-Saccharomyces yeasts towards indigenous yeast species of grape must, potential application in wine fermentation. J Appl Microbiol 2000; 89: 381-9. [ Links ]
44. Young TW. Killer yeasts, In: Rose AH, Harrison, JS, editors. The yeasts. London, United Kingdom, Academic Press, 1987, p. 131-64. [ Links ]
Recibido: 02/02/10
Aceptado: 23/09/10














