Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
Revista argentina de microbiología
versión impresa ISSN 0325-7541
Rev. argent. microbiol. vol.44 no.4 Ciudad Autónoma de Buenos Aires dic. 2012
AGENTES ANTIMICROBIANOS
First evaluation in Argentina of the GenoType® MTBDRplus assay for multidrug-resistant Mycobacterium tuberculosis detection from clinical isolates and specimens
Belén R. Imperiale1, Martín J. Zumárraga2, Gabriela Weltman3, Roxana Zudiker3, Ángel A. Cataldi2, Nora S. Morcillo1*
1Laboratorio de Referencia del Programa de Control de la Tuberculosis de la Provincia de Buenos Aires, Hospital Dr. Antonio Cetrángolo, Italia 1750, Florida (1602) Buenos Aires;
2Instituto Nacional de Tecnología Agropecuaria (CICV/INTA), Los Reseros and Nicolás Repetto, Hurlingham (1686) Buenos Aires; bioMèrieux, Arias 3751 (1430), Ciudad Autónoma de Buenos Aires, Argentina.
*Correspondence. E-mail: nora_morcillo@yahoo.com.ar
ABSTRACT
Tuberculosis (TB) and multidrug and extensively drug-resistant (DR) TB are important public health problems that are spreading worldwide. The aims of this study were to determine the sensitivity and specificity of the GenoType® MT BDRplus assay from smear-positive clinical specimens and isolates and to explore its possible application in routine work. Clinical samples were previously decontaminated using NaOH-N-acetyl-L-cystein or NaOH-ClNa hypertonic solution for Ziehl-Neelsen staining and cultures. The leftover sediments of smear-positive samples were stored at -20 ºC, 70 of which were selected to be included in this study according to their DR profile. Thirty DR Mycobacterium tuberculosis isolates were also assessed. Sequencing was used as gold standard to detect mutations conferring isoniazid (INH) and rifampicin (RIF) resistance. Valid results were obtained in 94.0 % of the samples and 85.5 % (53/62) of the INH-R samples were properly identified. Mutations in the katGS315T gene and inhA C-15T gene promoter region were present in 59.7 % (37/62) and 25.8% (16/62) of the INH-R samples, respectively. The system could also identify 97.7 % (41/42) of the RIF-R samples; the mutations found were rpoBS531L (66.7 %, 28/42), D516V (19.0 %, 8/42), H526Y and S531P/W (4.8 %, 2/42 each one), and S522L/Q (2.4 %, 1/42). A 98.8 % concordance between the GenoType assay and sequencing was obtained. GenoType® MT BDRplus has demonstrated to be easy to implement and to perform in clinical laboratories and useful for a rapid detection of DR M. tuberculosis from decontaminated sputa and clinical isolates. Therefore, this assay could be applied as a rapid tool to predict INH-R and/or RIF-R in DR risk cases.Key words: Molecular detection; Multidrug-resistant tuberculosis; Genotype® mtBdRplus.
RESUMEN
Primera evaluación en Argentina de GenoType® MTBDRplus para la detección de Mycobacterium tuberculosis multidrogo-resistente desde aislamientos y especímenes clínicos. La tuberculosis (TBC), y la TBC multi y extensivamente drogo-resistentes (DR) son importantes problemas de salud pública mundial. El objetivo de este estudio fue determinar la sensibilidad y especificidad del sistema GenoType® MT BDRplus a partir de esputos (baciloscopía positiva) y aislamientos clínicos de Mycobacterium tuberculosis, explorando su aplicación clínica. Previo a la tinción de Ziehl-Neelsen y al cultivo, las muestras fueron descontaminadas mediante NaOH-N-acetyl-L-cisteina o la solución hipertónica NaOH-NaCl. Los sedimentos remanentes se conservaron a -20 ºC, y 70 de ellos fueron incluidos en este estudio según su perfil de DR. Treinta cepas de M. tuberculosis DR fueron también evaluadas. La secuenciación fue utilizada como método de referencia para la detección de mutaciones que confieren resistencia a isoniacida (INH) y rifampicina (RIF). Se obtuvieron resultados válidos en el 94,0 % de las muestras, identificándose al 85,5 % (53/62) de las INH-R. La mutación katG S315T estuvo presente en 59,7 % (37/62), y la mutación C-15T del promotor del gen inhA en 25,8 % (16/62) de las mismas. El sistema identificó el 97,7 % (41/42) de las muestras RIF-R. Las mutaciones encontradas fueron rpoB S531L (66,7 %, 28/42), D516V (19,0 %, 8/42), H526Y y S531P/W (4,8 %, 2/42 cada una de ellas) y S522L/Q (2,4 %, 1/42). La concordancia entre el GenoType y la secuenciación fue del 98,8 %. El sistema GenoType® MT BDRplus resultó ser útil, fácil de realizar e implementar para la detección rápida de M. tuberculosis DR. Por lo tanto, este ensayo podría ser aplicado como una herramienta rápida para el diagnostico de TBC DR, principalmente en aquellos casos asociados a factores de riesgo.Palabras clave: Detección molecular; Tuberculosis multidrogo-resistente; Genotype® mtBdRplus.
INTRODUCTION
Tuberculosis (TB) and especially multidrugresistant TB (MD R-TB), TB resistant to isoniazid (INH) and rifampicin (RIF), and the most recently described extensively drug- resistant TB (XD R-TB) caused by MD R Mycobacterium tuberculosis, which is also resistant to kanamycin, amikacin or capreomycin and one fluoroquinolone, are important public health problems that are spreading worldwide (16, 17). Currently, the World Health Organization (WHO) estimates that approximately 500,000 new cases of MD R-TB occur each year (32), 5 to 7 % of which might evolve to XD R (29, 31). These alarming figures have highlighted the urgency for rapid screening methods to detect DR in a short turnaround time, especially in those patients with high risk of having MD R/XD RTB, such as potential institutional transmission and cases with HIV co-infection (30).
Argentina is a country with middle incidence TB rate (23.2 per 100,000 inhabitants in 2010), (14) being Buenos Aires the province having the highest number of cases, 4,298 and an incidence of 28.1 per 100,000 inhabitants. Moreover, 80 % of these cases are concentrated in the overcrowded suburban areas of Buenos Aires City (23).
According to the last national drug-resistance (DR) surveillance, it was observed that 15.5 % of patients with a previous anti-TB treatment history and 2.2 % of the new cases were caused by MD R strains and that the prevalence of INH resistance is about 17 % (26).
INH and RIF are two of the main anti-tuberculosis first line drugs used in the standard treatment regimes for TB. Resistance to INH is generally the first step for developing MD R-TB and resistance to RIF is commonly a marker of MD R-TB, since more than 90 % of the M. tuberculosis RIF-R strains are also resistant to INH. Furthermore, INH is currently used for chemoprophylaxis in children with a proven fully drug-susceptible TB contact.
When resistance to INH and RIF occurs with or without associated resistance to any other drug, the case is defined as a MD R-TB case (16, 17).
As it was previously described, DR in M. tuberculosis is mainly caused by point mutations in certain genes (3). A base-pair change in codon 315 of the katG gene is the most common mutation associated to INH-R, followed by mutations in position -15 of the inhA gene promoter region (4, 10, 13). Mutations within the hot spot region of the rpoB gene of M. tuberculosis, mainly in codons 516, 526 and 531 are responsible for about 95 to 97.0 % of RIF-R (5, 18, 21).
In our setting, detection of DR-TB is mainly based on phenotypic drug susceptibility testing (DST). Faster methods for rapid detection of DR-TB to avoid incorrect anti-TB treatment and to prevent transmission of resistant forms of the disease in the community are urgently needed.
The GenoType® MT BDRplus assay is a rapid molecular method that can be used from M. tuberculosis clinical isolates or directly from pulmonary smear-positive clinical specimens. This method has been designed to identify RIF-R by the detection of the most common mutations within the hot spot region of the rpoB gene. For the INH-R detection, the katG gene including codon 315 and the promoter region of the inhA gene, position -15 are examined.
The GenoType® MT BDRplus assay is based on DNA -Strip technology, and the whole procedure is divided into three steps: DNA extraction, a multiplex amplification with biotinylated primers, and a reverse hybridization. Hybridization includes the following steps: chemical denaturation of PCR products, hybridization of the single-stranded, biotin-labeled amplicons to membrane-bound probes, stringent washing, addition of a streptavidin/alkaline phosphatase conjugate and a conjugate-mediated reaction. A banding pattern is obtained with easy and fast interpretation of the results (7).
The aims of this study were: a) to determine the sensitivity and specificity of the GenoType® MTBDRplus assay directly applied from smearpositive clinical specimens and isolates, in comparison to conventional drug susceptibility testing (DST) methods and sequencing of katG, inhA (promoter region) and rpoB genes of M. tuberculosis; b) to explore its possible application in routine work and under clinical conditions.
MATERIALS AND METHODS
It was a retrospective study that included smear-positive clinical specimens and M. tuberculosis isolates obtained from patients diagnosed in the Reference Laboratory of Tuberculosis Control Program at Dr. Cetrangolo Hospital of Buenos Aires Province.
Clinical isolates. A total of 30 previously characterized DR strains of M. tuberculosis were assessed.
Clinical specimens. In order to obtain isolates, clinical samples were decontaminated by the NaOH-N-acetyl- L-cystein or by the hypertonic solution of NaCl following previously described protocols (20). The concentrated sediment was used for preparing smears for Ziehl-Neelsen staining (15) and for cultures in solid [Löwenstein-Jensen (LJ), and Stonebrink (Sk)], and liquid media BACTE C MG IT 960 system (BD, Buenos Aires, Argentina) (8).
The leftover sediments of smear-positive samples were stored at -20ºC to be used with the GenoType assay.
Drug-susceptibility testing to first anti-TB drugs were performed -from cultures and in all specimens- by the indirect proportion method on LJ, the BACTE C MG IT 960 SIRE kit (BD, Argentina) and in some cases by the microplate colorimetric method (2, 9, 19).
According to the drug resistance pattern obtained by the DST, 70 frozen leftover sediments of smear-positive samples were selected to be tested by the GenoType system.
Table 1 shows the total number of clinical specimens and isolates included in the study and their drug-resistant profiles.
Table 1. Samples included in the study and their drug-resistant profile
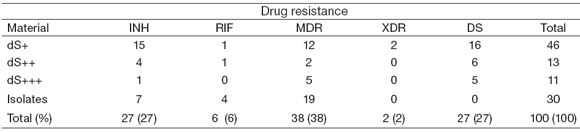
The pluses indicate bacillary load within the clinical specimen (+: about 1x104 bacilli per ml of sputum; ++: ≥ 1x105 bacilli/ml; +++: ≥ 1x106 bacilli/ml); dS: decontaminated sputa; INH: isoniazid; RIF: rifampicin; MDR: multidrug-resistant; XDR: extensively drug-resistant; DS: drugsusceptible.
The fully drug-susceptible (DS) reference strain M. tuberculosis H37Rv AT CC 29274 was used as control for DST.
M. tuberculosis identification
The molecular technique spoligotyping (27), which is specific for the differentiation of most of the members of the M. tuberculosis complex, was performed on clinical isolates (30 prior clinical isolates plus the 70 cultures grown from the decontaminated sputa tested) to confirm that they were all M. tuberculosis.
DNA sequencing
This technique was used as a gold standard molecular method to detect mutations conferring INH and RIF resistance. The M. tuberculosis clinical isolates and the cultures obtained from the decontaminated sputa included in the study were sequenced.
To characterize the mutations conferring INH and RIF-R in the samples included in the study, M. tuberculosis clinical isolates and cultures obtained from the decontaminated sputa were sequenced. To accomplish the above mentioned aim, a 435 bp fragment containing codon 315 of the katG gene, 648 bp of the inhA promoter (including position -15) and 250 bp of the flanking region of the "hot spot" rpoB gene were amplified and sequenced.
Both DNA strands were sequenced in all cases. H37Rv AT CC 27294 was used as wild type (WT) control for sequencing. Table 2 shows the primers used for PCR and sequencing of the studied genes. These primers and the PCR protocols used for sequencing were adopted from previous studies (13).
Table 2. Primers used for PCR and DNA sequencing of M. tuberculosis drug-resistant genes
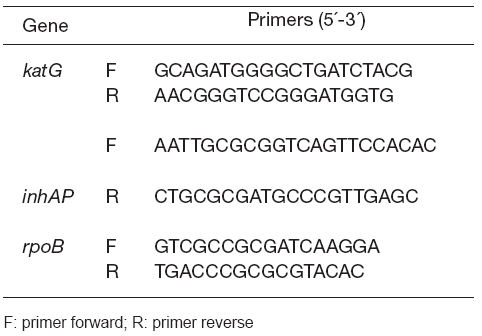
The MyCyclerTM thermal cycler (BioRad Richmond, CA, USA) was used to perform the PCR reactions. PCR products were purified by the QIAquick PCR purification kit (QIAGEN GmbH, Hilden, Germany) and quantified on a 0.8 % agarose gel (BioRAD ). The purified PCR products were later sequenced using a DNA sequencer 3130xl Genetic Analyzer (Applied Biosystems, Buenos Aires, Argentina). Double- stranded DNA was sequenced for each isolate in order to avoid discrepancies and the results were analyzed by the Basic Local Alignment search Tool (National Center for Biotechnology Information, Bethesda, MD, USA).
GenoType® MTBDRplus assay
The previously mentioned 100 samples were tested by the GenoType assay according to the manufacturer's instructions (Hain Lifescience).
In order to obtain DNA from the clinical specimens, the decontaminated sputa stored at -20ºC were thawed and 500 μl of each were centrifuged during 15 min at 10,000 x g, then the pellet was resuspended in 100 μl of molecular biology- grade water, heated at 95 ºC for 20 min, sonicated (LysorTM LCx Probe System, Abbott Laboratories) for 15 min and centrifuged for 5 min at 13,000 x g.
To obtain DNA from clinical isolates, 2 colonies from solid media were resuspended in 300 μl of molecular biology-grade water and heated at 95 ºC during 20 min, then the samples were centrifuged for 5 min at 13,000 x g.
The PCR mixture contained 5 μl of crude extracted DNA, 35 μl of a primer nucleotide mixture (provided by the kit), 5 μl of 10X PCR buffer containing 15 mM MgCl2, 2 μl of 25 mM MgCl2, 1 U hot start Taq polymerase (QIAGEN, GmbH, Hilden, Germany) and molecular-grade water to a final volume of 50 μl.
The thermal cycler parameters consisted of 15 min of initial hot start Taq polymerase activation and DNA denaturization at 95 ºC, followed by 10 cycles of 30 s at 95 ºC and 2 min at 58 ºC; then 20/30 cycles (isolates/clinical samples) of 25 s at 95 ºC, 40 s at 53 ºC and 40 s at 70 ºC; finally, 8 min at 70 ºC for elongation.
RESULTS
Molecular identification of the samples
All 100 specimens, both the 30 previously obtained isolates and the 70 sputa cultured as part of the routine laboratory procedures, were identified as M. tuberculosis by spoligotyping.
GenoType® MTBDRplus assay results
Valid results were obtained in 94 out of 100 (94 %) samples included in the study.
Six decontaminated smear-positive sputa classified as one plus (+, about 10,000 bacilli per ml of sputum) from 2 INH-R specimens, 1 RIF-R, 2 MD R and 1 XD R did not show any signals on the GenoType strip for any one of the three different genes.
Table 3 shows the mutations found by the GenoType assay for the samples that yielded valid results.
Table 3. Mutations found in clinical specimens and strains with GenoType®MTBDRplus assay
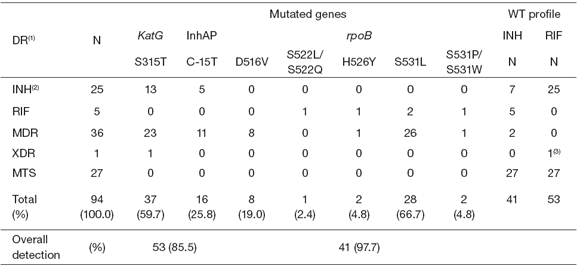
By using the GenoType assay, we could properly identify 85.5 % (53/62) of the INH-R samples detected by DST.
The mutation S315T in the katG gene was present
in 59.7 % (37/62) of the INH-R samples, whereas the mutation C-15T of the inhA gene promoter region was in 25.8 % (16/62) of the INH-R specimens.
In 2 INH-R samples, the GenoType assay detected an unknown mutation in katG315 but AG C315AGA mutation was later confirmed by sequencing.
Regarding the rpoB gene mutations, the GenoType system identified 97.7 % (41/42) of the RIF-R samples. The S531L was the main mutation found in the rpoB gene (66.7 %, 28/42) of the RIF-R samples; 19 % (8/42) presented the D516V mutation, mutations H526Y and S531P/W were both found in 4.8 % (2/42) each one, and the S522L/Q mutation was present only in one sample (2.4 %, 1/42).
All DS clinical specimens and H37Rv strains assessed in the study showed the WT sequence for all the fragments of the three genes included in the strips.
Being a retrospective study, the specificity was evaluated by taking into account the DS isolates from clinical specimens, the reference strain H37Rv and considering the DR patterns of the isolates. For instance: one resistant strain to RIF only should have wild type (WT) katG and inhA promoter genes, while a strain only resistant to INH should show a WT rpoB gene. No false positive signals were observed in any case, therefore the specificity of the GenoType assay in this study was considered to be 100 %.
Concordance between GenoType® MTBDRplus assay and DNA sequencing
DNA sequencing results were obtained in 84 out of the 100 samples studied by the GenoType assay.
A 98.8 % global concordance between the GenoType assay and DNA sequencing was obtained. When the mutation was present in C-15T of inhA or in katG315 the concordance between both methods was 100 %, but a 97.2 % concordance was obtained for detection of rpoB mutations: one sample showing M. tuberculosis XD R profile by DST carried the GA C516GT C rpoB mutation by sequencing, which was detected as WT by the GenoType assay.
The kappa (κ) coefficient for the concordance between both methods was calculated using the MedCalc® Software v 9.5.2.0 (Mariakerke, Belgium) and it was 0.996, indicating an excellent agreement between both methods (κ > 0.8) and a very good concordance (24). This misdetected sample by GenoType was confirmed as carrying the GA C516GT C mutation by sequencing it twice. The RIF-R detected by phenotypic DST methods was also confirmed by therapeutic evidence, based on patient response to anti-TB specific treatment.
DISCUSSION
In this study, the evaluation of the clinical performance of GenoType® MT BDRplus was carried out with the main objective of comparing its sensitivity and specificity to conventional DST and to explore its possible application in routine work under clinical conditions. The overall sensitivity of the GenoType assay obtained in this study was 94 % because 6 decontaminated sputum samples did not show any signals in the GenoType strips. The presence of inhibitors in the 6 clinical samples was discarded by â-actin gene amplification (25) and no GenoType PCR products from any of the samples were evidenced on an agarose gel. Therefore, a possible explanation for the negative GenoType results could be an insufficient amount of mycobacteria in those six samples. Since the global sensitivity of this kit is around 10,000 mycobacteria in 500 μl of decontaminated sputa used for DNA extraction, a loss of DNA during the extraction process or its relative inefficiency would explain, at least partially, the decreased sensitivity. Valid sequencing results for the different genes were obtained from all these samples. Nevertheless, global specificity and concordance were very good, accounting for 100 % and 98.8 %, respectively.
As previously reported by other authors, in this study the katG gene mutation S315T and the rpoB gene mutation S531L were the most prevalent mutations found to be responsible for INH-R and RIF-R, respectively (17, 18, 20, 21).
The GenoType® MT BDRplus assay was demonstrated to be easy to implement and perform in clinical laboratories and useful for a rapid detection of M. tuberculosis resistant to INH and RIF from both decontaminated sputa and clinical isolates. Therefore, this assay could be applied as a rapid screening tool to predict INH-R and RIF-R, especially in those cases at high risk of becoming infected by a DR or a MD R strain. The screening would also be useful in other situations such as HIV co-infection, immunosuppression or malignancies and both adult and children household contacts of MDR-TB cases.
This assay has been used worldwide (1, 6, 11, 12, 22, 24, 28) and it has been recently endorsed by WHO to be used as a molecular screening tool for rapid detection of MD R-TB cases (30).
However, it is worth noting that this test cannot totally replace the conventional phenotypic DST method in clinical practice (30) because DST is still necessary to confirm XD R-TB. As it was shown in this study, a few DR cases could not be detected by the Genotype assay. The most plausible explanation may reside in the fact that only the most common mutations conferring drugresistance are detected by the strip. Moreover, different drug-resistance mechanisms such as those implying efflux pumps might be involved in the occurrence of this phenomenon. Based on the sensitivity and specificity values found in this study, the method could be reliable with a result showing "resistance" to any given drug. Only those cases with invalid results (band pattern showing heteroresistance) or those designated as "susceptible" to a drug but being highly suspicious for drug-resistance should be confirmed by a phenotypic DST method. Consequently, only few cases should require confirmation after obtaining either drug-resistant or drug-susceptible results by the GenoType assay. The importance of adding a molecular test for drugresistance detection at a clinical level is also evident at the moment of considering rapidness and optimal assignment of economic and personnel resources. When the GenoType assay is applied as screening tool, there is proven time-saving in technicians' work and also a reduction in the costs of commercial DST methods, which together compensate for the cost of molecular techniques used as their replacement. The whole healthcare system could benefit from the implementation of rapid diagnostic tools, leading to a shorter hospitalization time, with all the implications brought about to the affected families by this particular socio-economic setting.
Acknowledgements: we especially thank Hain Lifescience for kindly providing us the GenoType® MT BDRplus kit assay. AC is a career member and BI is a fellow from CON ICET, Argentina.
1. Bazira J, Asiimwe BB, Joloba ML, Bwanga F, Matee MI. Use of the GenoType® MT BDRplus assay to assess drug resistance of Mycobacterium tuberculosis isolates from patients in rural Uganda. BMC Clin Pathol 2010; 10: 5. [ Links ]
2. BD BACTEC MGIT 960 SIRE kits for the antimycobacterial susceptibility testing of Mycobacterium tuberculosis. Becton Dickinson. Sparks, MD, USA: Becton Dickinson, 2005. [ Links ]
3. Bravo LT, Tuohy MJ, Ang C, Destura RV, Mendoza M, Procop GW, Gordon SM, Hall GS, Shrestha NK. Pyrosequencing for rapid detection of Mycobacterium tuberculosis resistance to rifampin, isoniazid and fluoroquinolones. J Clin Microbiol 2009; 47: 3985-90. [ Links ]
4. Cardoso RF, Cooksey RC, Morlock GP, Barco P, Cecon L, Forestiero F, Leite CQ, Sato DN , Shikama ML, Mamizuka EM, Hirata RD, Hirata MH. Screening and characterization of mutations in isoniazid-resistant Mycobacterium tuberculosis obtained in Brazil. Antimicrob Agents Chemother 2004; 48: 3373-81. [ Links ]
5. Caws M, Duy PM, Tho DQ, Lan NT, Hoa DV, Farrar J. Mutations prevalent among rifampin- and isoniazidresistant Mycobacterium tuberculosis isolates from a hospital in Vietnam. J Clin Microbiol 2006; 44: 2333-7. [ Links ]
6. Coronel J, Roper M, Mitchell S, Castillo E, Gamarra N, Drobniewski F, Luna G, Mendoza A, Moore DA. MODS accreditation process for regional reference laboratories in Peru: validation by GenoType® MT BDRplus. Int J Tuberc Lung Dis 2010; 14: 1475-80. [ Links ]
7. Hain Lifescience GmBH. GenoType®MTBDRplus kit manual, p. 23. [ Links ]
8. Hanna B, Ebrahimzadeh A, Elliott B, Morgan M, Novak SM, Rusch-Gerdes S, Acio M, Dunbar DF, Holmes TM, Rexer CH, Savthyakumar C, Vannier AM. Multicenter evaluation of the Bactec MG IT 960 system for the recovery of mycobacteria. J Clin Microbiol 1999; 37: 748-52. [ Links ]
9. Heifets LB. Conventional methods for antimicrobial susceptibility testing of M. tuberculosis. In: Bastian I, Portaels F, editors. Multidrug-resistant tuberculosis. London, UK, Kluwer Academic Publishers, 2001, p. 135-6. [ Links ]
10. Heym B, Saint-Joanis B, Cole ST. The molecular basis of isoniazid resistance in Mycobacterium tuberculosis. Tuber Lung Dis 1999; 74: 267-71. [ Links ]
11. Hillemann D, Rüsch-Gerdes S, Richter E. Evaluation of the GenoType MT BDRplus assay for rifampin and isoniazid susceptibility testing of Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol 2007; 8: 2635-40. [ Links ]
12. Huyen M, Tiemersma E, Lan N, Cobelens F, Dung N, Sy D, Buu T, Kremer K, Hang P, Caws M, O´Brien R, van Soolingen D. Validation of the GenoType® MT BDRplus assay for diagnosis of multidrug resistant tuberculosis in South Vietnam. BMC Infect Dis 2010; 10: 149. [ Links ]
13. Imperiale BR, Cataldi AA, Morcillo NS. Rapid detection of multidrug-resistant Mycobacterium tuberculosis by multiplex allele-specific polymerase chain reaction. Int J Tuberc Lung Dis 2011; 15: 496-501. [ Links ]
14. Instituto Nacional de Enfermedades Respiratorias Emilio Coni. Situación de la tuberculosis en la Argentina. Ministerio de Salud y Medio Ambiente de la Nación. Septiembre 2011. [ Links ]
15. Kent PT, Kubica GP. Public Health microbiology. A guide for a level III laboratory. Centers for Disease Control, Atlanta GA, 1985. [ Links ]
16. Mattelli A, Migliori GB, Cirillo D, Centis R, Girardi E, Raviglione M. Multidrug-resistant and extensively drugresistant Mycobacterium tuberculosis: epidemiology and control. Expert Rev Anti Infect Ther 2007; 5: 857-71. [ Links ]
17. Migliori GB, Loddenkemper R, Blasi F, Raviglione MC. 125 years after Robert Koch's discovery of the tubercle bacillus: the new XD R-TB threat. Is "science" enough to tackle the epidemic? Eur Respir J 2007; 29: 423-7. [ Links ]
18. Mokrousov I, Bhanu NV, Suffys PN, Kadival GV, Yap SF, Cho SN, Jordaan AM, Narvskava O, Singh UB, Gomes HM, Lee H, Kulkami SP, Lim KC, Khan BK, van Soolingen D, Victor TC, Schouls LM. Multicenter evaluation of reverse line blot assay for detection of drug resistance in Mycobacterium tuberculosis clinical isolates. J Microbiol Methods 2004; 57: 323-35. [ Links ]
19. Morcillo N, Imperiale B, Di Giulio B. Evaluation of MG IT 960TM and the colorimetric based method for tuberculosis drug susceptibility testing. Int J Tuberc Lung Dis 2010; 14: 1169-75. [ Links ]
20. Morcillo N, Imperiale B, Palomino JC. New simple decontamination method improves microscopic detection and culture of mycobacteria in clinical practice. J Infect Drug Resist 2008; 1: 21-6. www.ncbi.nlm.nih. gov/pmc/articles/PM C3108717/?tool=pubmed. [ Links ]
21. Morcillo N, Zumárraga M, Alito, Dolmann A, Schouls L, Cataldi A, Kremer K, van Soolingen D. A low cost, homemade, reverse-line blot hybridization assay for rapid detection of rifampicin resistance in Mycobacterium tuberculosis. Int J Tuberc Lung Dis 2002; 6: 959-65. [ Links ]
22. Nikolayevskyy V, Balabanova Y, Simak T, Malamanova N, Fedorin I, Drobniewski F. Performance of the GenoType MT BDRplus assay in the diagnosis of tuberculosis and drug resistance in Samara, Russian Federation. BMC Clin Pathol 2009; 9: 2. [ Links ]
23. Programa de Control de Tuberculosis de la Provincia de Buenos Aires. Informe Epidemiológico Año 2010. Ministerio de Salud, La Plata. Agosto de 2012. [ Links ]
24. Rigouts L, Hoza AS, De Rijk P, Torrea G, Chonde TM , Basra Z, Zignol M, van Leth F, Egwaga SM, van Deun A. Evaluation of the GenoType® MT BDRplus assay as a tool for drug resistance surveys. Int J Tuberc Lung Dis 2011; 15: 959-65. [ Links ]
25. Selvey S, Thompson EW, Matthaei K, Lea RA, Irving MG, Griffiths LR. ß-actin unsuitable internal control for RTPCR. Mol Cell Probes 2001; 15: 307-11. [ Links ]
26. Servicio Micobacterias. Instituto Nacional de Enfermedades Infecciosas- ANLIS "Dr. Carlos G. Malbran", Argentina. XL Reunión Nacional Anual del Consejo Confederal de Tuberculosis. Buenos Aires, 24 y 25 de noviembre de 2010. [ Links ]
27. van Embden JD, van Gorkom T, Kremer K, Cansen R, van der Zeijst BA, Schouls LM. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J Bacteriol 2000; 182: 2393-401. [ Links ]
28. Vijdea R, Stegger M, Sosnovskaja A, Andersen AB, Thomsen VØ, Bang D. Multidrug-resistant tuberculosis: rapid detection of resistance to rifampin and high or low levels of isoniazid in clinical specimens and isolates. Eur J Clin Microbiol Infect Dis 2008; 27: 1079-86. [ Links ]
29. WHO. Anti-tuberculosis drug resistance in the world. Fourth global report. 2008. WHO/HTM/TB/2008.394. [ Links ]
30. WHO. Molecular line probes assays for rapid screening of patients at risk of multidrug-resistant tuberculosis. http://www.who.int/tb/features_archive/policy_statement.pdf. June 2008. [ Links ]
31. WHO. Multidrug and extensively drug-resistant TB (M/XD R-TB): 2010 global report on surveillance and response, 2010. WHO/HTM /TB/2010.3. ted May 2011. [ Links ]
32. WHO/IUATLD. Report 3, Geneva. WHO, 2004, p. 146-7. [ Links ]
Recibido: 8/5/2012
Aceptado: 6/6/2012














