Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
Revista argentina de microbiología
Print version ISSN 0325-7541On-line version ISSN 1851-7617
Rev. argent. microbiol. vol.48 no.2 Ciudad Autónoma de Buenos Aires June 2016
http://dx.doi.org/10.1016/j.ram.2015.12.003
BRIEF REPORTS
http://dx.doi.org/10.1016/j.ram.2015.12.003
Arcobacter butzleri survives within trophozoite of Acanthamoeba castellanii
Arcobacter butzleri sobrevive en el interior de trofozoitos de Acanthamoeba castellanii
María P. Villanuevaa, Gustavo Medinab, Heriberto Fernándeza, *
a. Instituto de Microbiología Clínica, Universidad Austral de Chile, PO Box 567, Valdivia, Chile
b. Facultad de Ciencias de la Salud, Universidad Católica de Temuco, PO Box 15-D, Temuco, Chile
Received 2 March 2015; accepted 1 December 2015
Available online 11 March 2016
* Corresponding author. E-mail address: hfernand@uch.cl (H. Fernández).
0325-7541/© 2016 Asociacón Argentina de Microbiología. Published by Elsevier España, S.L.U. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Abstract
The survival of three Arcobacter butzleri strains inside Acanthamoeba castellanii was assessed using axenic cultures of A. castellanii that were inoculated with the tested strains and incubated at 26 °C under aerobic conditions for 240 h. The behavior of bacteria in contact with amoebae was monitored using phase contrast microscopy. The bacterial survival rate within amoebae was assessed through counting colony forming units, using the gentamicin protection assay. All A. butzleri strains were able to survive during 240 h within the amoebae, thus suggesting that (i) A. butzleri resists the amoebic digestion processes at least for the analyzed time; (ii) that A. castellanii could serve as an environmental reservoir for this bacterium, probably acting as a transmission vehicle for A. butzleri.
Keywords
Arcobacter butzleri; Acanthamoeba castellanii; Survival; Environmental reservoir.
Resumen Se determinó la sobrevida de 3 cepas de Arcobacter butzleri en el interior de trofozoítos de Acanthamoeba castellanii utilizando cultivos axénicos de la ameba, los que fueron inoculados con las cepas bacterianas e incubados a 26 °C en aerobiosis durante 240 h. La interacción bacterias/amebas fue monitorizada mediante microscopía de contraste de fase. Las tasas de sobrevida bacteriana en el interior de las amebas fueron establecidas a través del recuento de unidades formadoras de colonias mediante ensayos de protección con gentamicina. Todas las cepas de A. butzleri fueron capaces de sobrevivir en el interior de la ameba al menos durante 240 h, lo que sugiere, por un lado, que A. butzleri puede resistir los procesos de digestión propios de la ameba, al menos por el lapso aquí analizado y, por el otro, que A. castellanii podría actuar como reservorio ambiental de esta bacteria y, probablemente, como vehículo de transmisión de A. butzleri.
Palabras clave
Arcobacter butzleri; Acanthamoeba castellanii; Sobrevida; Reservorio ambiental.
The genus Arcobacter was originally proposed by Vandamme et al.14 and includes bacteria previously considered as members of the genus Campylobacter. Today, the genus Arcobacter is represented by 21 species isolated from human, animals and environmental samples15. However, only four species have been isolated from human and animal samples and they are considered of medical interest: Arcobacter cryaerophilus, Arcobacter butzleri, Arcobacter skirrowii and Arcobacter thereius13.
A. butzleri is the species that is most frequently isolated from environmental water, food and clinical samples. From a public health point of view, it is considered an emerging foodborne and zoonotic enteropathogen transmitted by food and water. Furthermore, it has been ranked as the fourth most common member of Campylobacteraceae isolated from human feces2,5,13. Thus, it has become increasingly important as an agent of diarrhea in human beings, recognizing as potential routes of infection the consumption and manipulation of contaminated raw or poorly cooked food of animal origin and untreated water2,5,13.
One of the main reservoirs of this species is water in diverse environments2 where they may interact with other microorganisms, which are natural inhabitants of hydric ecosystems, in particular with free-living amoebas.
Free-living amoebas of the Genus Acanthamoeba are widely distributed in water environments. Amoebic trophozoites are part of the indigenous inhabitants of natural water bodies, sharing this habitat with other microorganisms, mainly bacteria. Since these bacteria can survive for diverse periods within the amoeba, many researchers have suggested that these protozoa may be a reservoir for bacterial species1. Because of the particular ability of some bacteria to survive within amoebae, the members of genus Acanthamoeba could play a role in the transmission of some enteric bacteria. The survival of Campylobacter jejuni inside Acanthamoeba and its transmission to chicken as an endosymbiont of A. castellanii and Acanthamoeba polyphaga1 has been demonstrated.
Since A. butzleri is a bacterium that can be recovered from water environments and food products, such as mussels produced in estuarine environments2,6, it could share this environment with free-living amoebas, probably establishing close relationships with these protozoa, similarly to those described in the genus Campylobacter1,11,12.
In order to demonstrate if A. butzleri is capable of establishing similar close relations with free-living amoebas, we determined the survival capacity of Arcobacter inside A. castellanii.
Three strains of A. butzleri were used for this study. Strain F215 (isolated from child human stools), strain Puar190 (isolated from chicken meat) and the reference strain LMG10828 (isolated from human feces) used as control strain and kindly provided by Dr. Peter Vandamme (Laboratory of Microbiology, Ghent University, Belgium). These strains were stored at −80 °C in brain-heart infusion broth supplemented with 15 % glycerol. Before each experiment, bacteria were grown on blood agar plates incubated at 26 °C for 48 h under aerobic conditions.
A. castellanii strain BP91/2760 was originally isolated from the eye of a patient with keratitis and was kindly provided by Dr. Winiecka-Krusnell (Public Health Agency of Sweden). Amoebae were maintained axenically in peptone yeast extract glucose (PYG) medium at 26 °C as monolayers in 25 cm2 flasks and were examined under an inverted microscope before use. To assess the number of amoebae, counts were made using a Neubauer chamber.
To establish A. butzleri/A. castellanii early relationships, A. castellanii were grown to confluence in 25 cm2 flasks with 5 ml of PYG medium. The flask were gently shaken and the PYG with non-adherent amoebae was removed and replaced with fresh medium; then the amoebae were incubated on ice for 30 min to weaken adherence. The suspensions were centrifuged at 203 g for 10 min and the pellet was washed three times with Page's saline solution. After the last centrifugation, the pellet was suspended in PYG and the total number of amoebae was estimated. Aliquots of 1 ml with approximately 6 × 104 trophozoites were placed in each well of a 24-well tissue culture dish. Bacterial strains suspended in Page's saline solution to a density of approximately 1.2 × 108 colony forming units (CFU), and 100 ml of this suspension was added to each well containing the amoebae and the plate was incubated at 26 °C under aerobic conditions to induce the infection of the amoebas by Arcobacter strains in a 1:2000 ratio. As bacterial survival controls, three wells were inoculated with A. butzleri suspended in Page's saline solution without amoebae. Finally, the assessment of the early interactions between bacteria and amoebae was performed by taking aliquots of the suspension at 30 and 50 min and observed directly under phase contrast microscope.
Long-term interaction between bacteria and amoebae was studied at 24, 48, 72, 120, 168, and 240 h using a modification of the gentamicin protection assay proposed by Dirks and Quinlan3. In brief, after the early interaction assay, 100 μg/ml gentamicin were added to each well and incubated at room temperature for 1 h to kill extra-amoebic bacterial cells. Amoebas were harvested from the well and washed three times with Page's saline solution. After the third wash, an aliquot of 100 μl was taken and plated on blood agar plates to determine the number of extracellular bacteria that might have escaped the gentamicin treatment at each sampling time. 100 μl of 0.5% sodium desoxycholate was added to another aliquot for 25-30 min to perform the amoeba lysis. An aliquot of 100 μl of the lysate suspension was taken to count the number of total bacteria by CFU counts (intracellular + extracellular bacteria). Abundance of viable intracellular bacteria was determined by subtracting the number of extracellular bacteria from the total count. All these experiments were carried out in triplicate. The amoebae/bacteria interaction was also monitored and documented through phase contrast microscopy and by bright-field microscopy.
The results of the assays were divided into two groups in accordance with the duration of the tests performed.
A. - Early interactions between A. butzleri and A. castellanii. In the first 30 min of co-incubation, A. butzleri could be seen in close association with A. castellanii, at this time most of the bacteria showed a tendency to gather at one pole of the amoeba cell (Fig. 1A). Soon after 50 min of co-culture, A. butzleri cells were detected inside the amoeba. The internalized A. butzleri were located in vacuoles and never in the amoeba cytoplasm (Fig. 1B). These observations are consistent with our previous studies7,8, confirming an initial contact pattern in the interaction A. butzleri/A. castellanii.

Figure 1. Phase contrast microscopy. (A) Trophozoite of A. castellanii showing A. butzleri seen in close association with A. castellanii gathering at one pole of the amoeba cell known as food-cup (30 min of co-incubation). (B) Trophozoite of A. castellanii showing A. butzleri within the amoebic vacuoles (50 min of co-incubation).
The internalization of A. butzleri seems to be an active and probable metabolic process since all intracellular bacteria are located in vacuoles and never inside the amoeba cytoplasmic region. These images are similar to those previously reported by our group7,8.
This early observation indicated that the interaction between bacteria and amoebae could be a common phenomenon in Arcobacter, which is time-efficient. Recently, Medina et al.8 have reported that the attachment process of A. butzleri to A. castellanii involves the participation of mannose-binding proteins and membrane-associated receptors of glucose and galactose present in the amoebae, whereas in their internalization, the protozoan actin polymerization plays an active role.
B. - Time course of the interaction of A. butzleri and A. castellanii. The long-term experiments demonstrated that A. butzleri strains F-215 and PUAr190 were able to survive for at least 240 h inside the amoebas. On the other hand, control assays done to establish whether the presence of A. butzleri could affect the survival of the amoebae, showed no significant differences in the survival rates of the amoebae with and without bacteria (data not shown).
Regarding the survival of A. butzleri within A. castellanii trophozoites at different time points, all the bacteria studied in the co-culture with the amoeba as well as control bacteria showed a decrease in CFU values until reaching the minimum count on the tenth day. There is a clear difference in the survival of these three strains of A. butzleri inside the amoebae. The data suggest that recently isolated A. butzleri strains F-215 and PUAr190 have better survival than the reference strain LMG10828. Their colony counts suggest that this strain does not multiply inside A. castellani, not reaching the high numbers seen for the other two strains (Fig. 2). Furthermore, their UFC numbers drop in a similar way as the number of viable bacterial cells decays in the control assay without amoebae. Whether those differences are related to the source of isolation, geographic origin or number of subcultures is unknown, requiring further investigations.
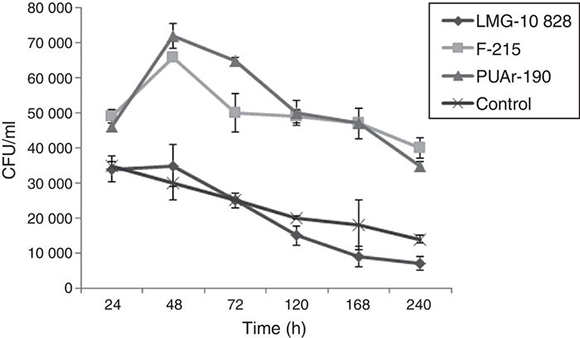
Figure 2. Average colony-forming units (CFU) of intracellular A. butzleri recovered after co-culture with A. castellanii. LMG-10828 control strain.
The methodology used in this study to estimate the intra-amoeba bacterial survival rate was selected because other methods, such as vital stains and counting by microscopy, express their results as percentages and do not allow to determine the bacterial number as accurately as the gentamicin protection assay does3.
The A. butzleri strains used in this study were not only able to infect A. castellanii but also capable of surviving inside the vacuoles, in which we confirmed that bacteria were actively motile. Through the phagolysosome formation analysis, Medina et al.8 concluded that the survival of A. butzleri inside the amoebae could be related to their ability to remain inside vacuoles not fused with lysosomes, or with their ability to retard phagolysosome formation. However, further investigations are needed to elucidate the survival mechanisms of A. butzleri inside free-living amoebae.
Transmissibility of enteropathogens, particularly those associated in zoonosis, has been difficult to identify and for the same reason, nearly impossible to prevent. In the last quarter of the 20th century and the beginning of this century, emerging pathogens have become a major health problem4. Recent observations showed that free-living amoebae may act as a reservoir and vehicle for the survival and transmission of C. jejuni and other enteropathogens, providing a unique form of self-perpetuation of these pathogens1,9,10,12.
Our team has been very interested in the role of Arcobacter species as human pathogens and its association with animal sources5,6. Different studies have indicated that A. butzleri can survive under several environmental conditions and can be isolated from diverse hosts like farm animals, seafood, and hose pets2,5,6. In the present study, the survival of A. butzleri inside A. castellanii strongly suggests that this protozoon can serve as a reservoir for A. butzleri and it might be an important vehicle in the transmission of this human emergent zoonotic pathogen, acting as a Trojan horse2,8,12. Experimental transmissibility assays of A. butzleri inside A. castellanii into mice, currently performed in our laboratory (data not shown), allow to infer the eventual participation of this protozoon as transmission vehicle of A. butzleri. Further studies have to be carried out to elucidate this epidemiological angle of the A. butzleri/free-living amoebae relationships.
Ethical disclosures
Protection of human and animal subjects
The authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of data
The authors declare that no patient data appear in this article.
Right to privacy and informed consent
The authors declare that no patient data appear in this article.
Financial support
Grants DID-UACH S-2007-37 and FONDECYT 1110202.
Conflict of interests
The authors declare that they have no conflict of interests.
Acknowledgements
To Professor Ph.D. Jadwiga Winiecka-Krusnell (Public Health Agency of Sweden, Sweden) for providing the A. castellanii T4 genotype strain; Professor Ph.D. Peter Vandamme (Ghent University, Belgium) for providing the A. butzleri reference strain LMG 10828; and Professor Ph.D. Guillermo Pérez-Pérez (New York University Langone Medical Center, U.S.A.) for their scientific advice and critical review.
1. Axelsson-Olsson D, Waldenström J, Broman T, Olsen B, Holmberg M. Protozoan Acanthamoeba polyphaga as a potential reservoir for Campylobacter jejuni. Appl Environ Microbiol. 2005;71:987-92. [ Links ]
2. Collado L, Figueras MJ. Taxonomy, epidemiology and clinical relevance of the genus Arcobacter. Clin Microbiol Rev. 2011;24:174-92. [ Links ]
3. Dirks BP, Quinlan JJ. Development of a modified gentamicin protection assay to investigate the interaction between Campylobacter jejuni and Acanthamoeba castellanii ATCC 30010. Exp Parasitol. 2014;140:39-43. [ Links ]
4. Duffy G, Lynch OA, Cagney C. Tracking emerging zoonotic pathogens from farm to fork. Meat Sci. 2008;78:34-42. [ Links ]
5. Fernández H, Flores S, Inzunza F. Arcobacter butzleri strains isolated from different sources display adhesive capacity to epithelial cells in vitro. Acta Sci Vet. 2010;38:287-91. [ Links ]
6. Fernández H, Villanueva MP, Mansilla I, Gonzalez M, Latif F. Arcobacter butzleri and A. cryaerophilus in human, animals and food sources, in southern Chile. Braz J Microbiol. 2015;46:147-57. [ Links ]
7. Fernández H, Villanueva MP, Medina G. Endosymbiosis of Arcobacter butzleri in Acanthamoeba castellanii. Rev Argent Microbiol. 2012;44:133. [ Links ]
8. Medina G, Flores-Martin S, Fonseca B, Otth C, Fernandez H. Mechanisms associated with phagocytosis of Arcobacter butzleri by Acanthamoeba castellanii. Parasitol Res. 2014;113: 1933-42. [ Links ]
9. Olofsson J, Axelsson-Olsson D, Brudin L, Olsen B, Ellström P. Campylobacter jejuni actively invades the amoeba Acanthamoeba polyphaga and survives within non-digestive vacuoles. PLoS ONE. 2013;8:e78873, http://dx.doi.org/10.1371/journal.pone.0078873. [ Links ]
10. Siddiqui R, Khan NA. Biology and pathogenesis of Acanthamoeba. Parasit Vectors. 2012;5:1-13. [ Links ]
11. Snelling W, Stern N, Lowery C, Moore J, Gibbons E, Baker C, Dooley J. Colonization of broilers by Campylobacter jejuni internalized within Acanthamoeba castellani. Arch Microbiol. 2008;189:175-9. [ Links ]
12. Vaerewijck MJM, Baré J, Lambrecht E, Sabbe K, Houf K. Interactions of foodborne pathogens with free-living protozoa: potential consequences for food safety. Compr Rev Food Sci. 2014;13:924-44. [ Links ]
13. Van den Abeele AM, Vogelaers D, Van Hende J, Houf K. Prevalence of Arcobacter species among humans, Belgium, 2008-2013. Emerg Infect Dis. 2014;20:1731-4. [ Links ]
14. Vandamme P, Vancanneyt M, Pot B, Mels L, Hoste B, Dewettinck D, Vlaes L, Van den Borre C, Higgins R, Hommez J, Kersters K, Butzler JP, Goossens H. Polyphasic taxonomic study of the emended genus Arcobacter with Arcobacter butzleri comb. nov. and Arcobacter skiwowii sp. nov., an aerotolerant bacterium isolated from veterinary specimens. Int J Syst Bacteriol. 1992;42:344-56. [ Links ]
15. Whiteduck-Léveillée K, Whiteduck-Léveillée J, Clotier M, Tambong JT, Xu R, Topp E, Arts MT, Chao J, Adam Z, Lévesque CA, Lapen DR, Villemur R, Talbot G, Khan IUH. Arcobacter lanthieri sp. nov. isolated from pig and dairy cattle manure. Int J Syst Evol Microbiol. 2015;65:2709-16. [ Links ]














