Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
Revista argentina de microbiología
versión impresa ISSN 0325-7541versión On-line ISSN 1851-7617
Rev. argent. microbiol. vol.51 no.3 Ciudad Autónoma de Buenos Aires set. 2019
http://dx.doi.org/10.1016/j.ram.2018.07.004
ORIGINAL ARTICLE
https://doi.org/10.1016/j.ram.2018.07.004
Genetic diversity of Mycoplasma hyopneumoniae in Mendoza province
Diversidad genética de Mycoplasma hyopneumoniae en la provincia de Mendoza
Camila Sosaa, Ariel Bloisb, Fernando Ibanezc,d, Pablo Tamiozzoa,*
a Departamento de Patología Animal, Facultad de Agronomía y Veterinaria, Universidad Nacional de Río Cuarto, Córdoba, Argentina
b Dirección Provincial de Ganadería, Ministerio de Economía, Infraestructura y Energía, Gobierno de Mendoza, Mendoza, Argentina
c Departamento de Ciencias Naturales, Facultad de Ciencias Exactas, Físico-Químicas y Naturales, Universidad Nacional de Río Cuarto, Córdoba, Argentina
d CONICET, Argentina
Received 12 July 2017; accepted 15 July 2018
Available online 14 January 2019
*Corresponding author.
E-mail address: ptamiozzo@ayv.unrc.edu.ar (P. Tamiozzo).
0325-7541/© 2018 Asociación Argentina de Microbiología. Published by Elsevier España, S.L.U. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Abstract In Argentina, enzootic pneumonia (EP) is highly prevalent and different genetic types of Mycoplasma hyopneumoniae have been identified. However, there is a lack of information about prevalence and other epidemiological aspects of EP in Mendoza province. A multiple Locus variable-number tandem repeat analysis (MLVA) targeting P97 R1, P97 R1A and P146 R3 loci was used to assess the genetic diversity of M. hyopneumoniae from clinical specimens recovered from pigs from five herds located in different districts of Mendoza province. M. hyopneumoniae could be typed from 27 bronchoalveolar lavages (BAL) specimens, and eight different MLVA types were identified. This is the first report about diversity of M. hyopneumoniae in Mendoza. Results obtained in this work allow drawing a better picture of the genetic diversity of this pathogen in Argentina.
KEYWORDS
Swine; Enzootic pneumonia; Mycoplasma hyopneumoniae; Genotypes; MLVA; Mendoza; Argentina
Resumen En Argentina, la neumonía enzoótica porcina (NEP) es altamente prevalente y se han identificado diferentes tipos genéticos de Mycoplasma hyopneumoniae. Sin embargo, se carece de información acerca de la prevalencia de NEP y de otros aspectos epidemiológicos de esta entidad en la provincia de Mendoza. En esta investigación se usó un análisis multilocus de regiones repetidas en tándem (MLVA) de los loci P97 R1, P97 R1A y P146 R3 para evaluar la diversidad genética de M. hyopneumoniae a partir de muestras clínicas de cerdos de cinco granjas localizadas en diferentes distritos de la provincia de Mendoza. M. hyopneumoniae pudo ser tipificado a partir de 27 muestras de lavado broncoalveolar (LBA) y se identificaron 8 diferentes MLVA-tipos. Este es el primer informe acerca de la diversidad genética de M. hyopneumoniae en Mendoza. Los resultados obtenidos permiten describir de manera más acabada la diversidad genética de este agente en nuestro país.
PALABRAS CLAVE
Cerdo; Neumonía enzoótica porcina; Mycoplasmahyopneumoniae; Genotipos; MLVA; Mendoza; Argentina
Introduction
Mycoplasma hyopneumoniae is the primary agent involved in porcine enzootic pneumonia (EP) M. hyopneumoniae infections are highly prevalent in almost all swine-producing areas, causing significant economic losses to the pig industry worldwide21.
In Argentina, pig production is mainly concentrated in Santa Fe, Buenos Aires and Córdoba. provinces. EP is highly prevalent both, in indoor and outdoor systems in Argentina5. However, there is a lack of information about prevalence and other epidemiological aspects of EP in other provinces in which pig production is less developed, as in Mendoza province.
Different genetic types of M. hyopneumoniae have been identified in herds from Córdoba, Santa Fe and San Luis provinces12,18,20; however they have never been reported in Mendoza. Thus, the objective of this study (n=100) was to assess the genetic diversity of M. hyopneumoniae circulating in herds from the province of Mendoza.
Material and methods
The study was performed according to the international guidelines of the Council for International Organizations of Medical Sciences (CIOMS).
Herds and study design
A cross-sectional study was carried out in an abattoir by collecting 20 broncho-alveolar lavage (BAL) specimens recovered from pigs from each of the five herds under study. Herds were located in different districts of Mendoza province. The main characteristics of the herds such as the kind of system and size (number of sows), among others are shown in Table 1.
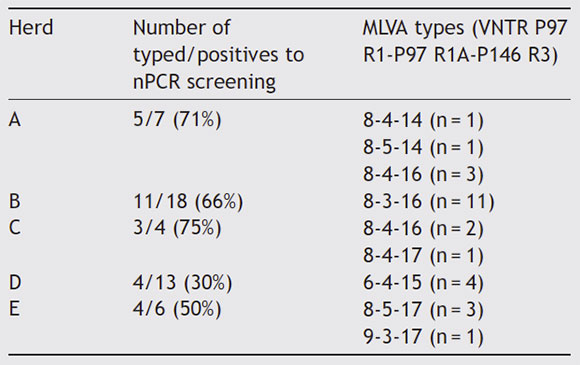
Table 1 ID of the herds, kind of production system (indoor, outdoor), size (number of sows), localization and status of vaccination against EP. Number of PCR positives out of samples (percentage) for M. hyopneumoniae detection from BAL specimens, according to the herd

Sample processing and M. hyopneumoniae typing
DNA from LBA specimens was extracted using DNAzol (Thermo Fisher Scientific, Argentina) according to the manufacturer's instructions. For M. hyopneumoniae detection 100 samples were analyzed by a nested-PCR1, considered as the screening technique.
In order to type M. hyopneumoniae, P97 R1, P97 R1A and P146 R3 loci were analyzed only in those specimens that were positive (48) to the nPCR screening mentioned above. Tandem repeat motives of P97 R1, P97 R1A and P146 R3 were amplified by a nested PCR (nPCR) format previously described13. After amplification, amplicons were purified (Puriprep-GP Kit, Inbio Highway, Argentina), quantified and sequenced (ABI 3130xl; Applied Biosystems, US) using the primers reported by Vranckx etal.22, for P97 and by Mayor et al.9, for P146. The number of tandem repeats was determined by analyzing the sequences with BioEdit 7.1.3.0 software7. For further analysis, only those specimens which were positive for both loci were considered.
Data analysis
A dendrogram based on the categorical values of the number of tandem repeats was constructed with the FAMD 1.20 software15 using the Dice similarity coefficient and the unweighted pair group method with arithmetic means (UPGMA). The number of tandem repeats from the 232, J, 7448, PMS and 7422 reference M. hyopneumoniae strains3 was included.
The Simpson's index of diversity of the combined variable-number of tandem repeat (VNTR) regions (P97 R1, P97 R1A and P146 R3) was calculated by using the HunterGaston diversity index - HDGI8.
Results
M. hyopneumoniae was detected by nPCR in all the herds in 48% (48/100) of the specimens. For P97, 27 specimens were positive and could be further sequenced and typed. Nested-PCR for P146 R3 showed more sensitivity, amplifying all processed BAL specimens. Thus, 27/48 (56%) of the specimens could be typed for both loci. The remaining specimens (n =21) could be typed only for P146 R3 and were not included in the analysis. Eight MLVA types were identified: 8-3-16 (n = 11), all of them obtained from herd B, 8-4-16 (n = 5) from herds A and C, 6-4-15 (n=4) from herd D, 8-5-17 (n = 3) from herd E, and the less frequent types: 84-14 (n = 1), 8-5-14 (n = 1), 8-4-17 (n = 1), 9-3-17 (n = 1) from herds A, C and E, respectively (Table 2). The numbers of tandem repeats of the analyzed loci for M. hyopneumoniae reference strains were: 13-1-21 (strain 232), 9-3-18 (strain J), 10-4-44 (strain 7448), 15-1-19 (strain PMS) and 12-3-40 (strain 7422).
Table 2 Number of typed samples out of positives to nPCR screening samples (percentage) and MLVA types (number of tandem repeats for P97 R1, P97 R1A and P146 R3) according to the herd
At a similarity cutoff of 85%, 13 clusters were observed -I to XIII - (Fig. 1). Most local genotypes were more related to other local genotypes than to the reference strains, with two exceptions (reference strain 7448 and local variant 64). Reference strain 7448 was more closely related to the local strains than to other reference strains. On the other hand, genetic variant 64 (obtained from herd E) was closely related to the reference strains than to the other native genotypes. Only one genetic variant of the agent was found in herd B (grouped into Cluster VII). This genotype was exclusively found in this herd. The same situation was observed in herd D (one genetic variant exclusively found in this herd, grouped into Cluster II). In herd A, three different genetic variants were observed, grouped in Clusters IV, V (variants exclusively found in this herd) and VIII (together with genotypes found in herd C). In herd C, two different variants were found. One genetic variant was exclusively found in this herd (Cluster III). The other variant corresponds to a genotype also found in herd A and grouped into Cluster VIII. Two different genotypes were observed in herd E. These genotypes were exclusively found in this herd and grouped into clusters VI and XI. All reference strains clustered into different groups (I, IX, XXII and XIII) (Fig. 1).
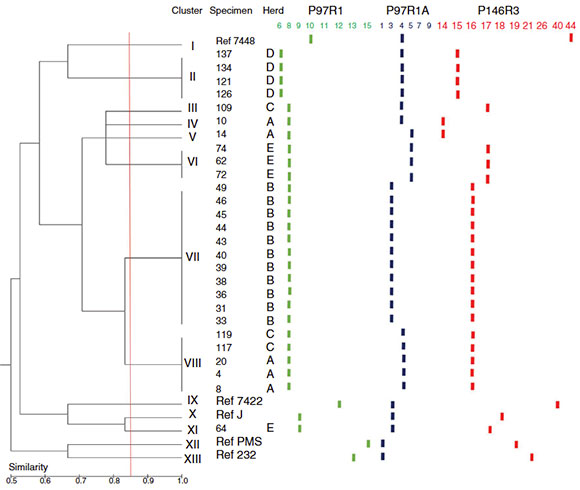
Figure 1 Dendrogram constructed with FAMD 1.20 software 15 using Dice similarity coefficient and unweighted pair group method with arithmetic means (UPGMA). Thirteen clusters (I to XIII) were identified at a similarity cutoff of 85%.
The number of tandem repeats for each locus and their combination are shown in Table 2. Simpson's index of diversity was D = 0.789.
Discussion
The circulation of different genetic types of M. hyopneumoniae in Mendoza province was demonstrated using a P97-P146 MLVA scheme. Since no molecular marker has been able to discriminate between high and low virulence M. hyopneumoniae strains, knowing the circulation of the different genotypes is important for the development of control strategies against EP. M. hyopneumoniae genetic diversity has been reported around the world at regional and herd level, between related and unrelated herds6,9,10,14,16,17,19.
The 2-loci (3-tandem repeat regions) MLVA profiles detected in this study can only be compared to the profiles reported by few studies, since neither these two loci were analyzed nor the number of repeat units present was determined in most reports. In this regard, some P97 R1 and P146 R3 MLVA types found here, were similar to those reported by Dos Santos et al. 6, in specimens from US and Spain. However, they did not consider the P97 R1A region, whose analysis increased the polymorphism. In Argentina, only one study used a similar MLVA Scheme12.
The discriminatory power was lower than the one reported by Dos Santos et al.6, mainly due to the number of processed specimens. It is worth mentioning that if the R1A region of P97 had not been included into the analysis, it would have obtained 5 MLVA types with a discriminatory power of 0.62 (data not shown).
Some studies reported the repeat number only for the P146 R3 motif, and they found similar polyserine repeats in some European countries9,14. Moreover, other studies reported the diversity of this gene, and some of them included P97, but did not determine the number of repeats2,4,10,11,22.
In Argentina there are previous reports about genetic diversity of this pathogen. However, none of the previous studies analyzed P97 R118,20. With regard to P146 R3, similar polyserine repeat motives were found in herds from Santa Fé, Córdoba, San Luis and in pigs from abroad18.
From the dendrogram analysis, it can be inferred that, in general, the genotypes from Mendoza province were more related to other native genotypes than to the reference strains. This observation agrees with a previous study in which the genetic difference between local M. hyopneumoniae types and bacterin strains was found20. More in-depth studies are necessary to determine the implications of such differences.
Only one genetic variant of the pathogen was found in herd B, being exclusive of this herd. The same situation was observed in herd D. These variants could represent well established M. hyopneumoniae strains and adapted to the conditions found in these herds, which agrees with previous results that showed persistence of mainly one distinct M. hyopneumoniae type within animals of the same herd23. However, the circulation of other less frequent types of the agent into the farms cannot be excluded. Different genetic variants were found in herds A, C and E. Such diversity observed in these herds could be explained by the access of different genetic types through replacement animals, wild pigs, personnel, fomites or airborne transmission. Regrettably, this information was not taken into account in this study.
As we previously proposed12 and considering the difficulty in comparing the results from different studies, we consider that the use of a standardized nomenclature for MLVA loci and alleles of M. hyopneumoniae is highly needed. Furthermore, the development of an online database including MLVA profiles for this pathogen would be of great help to share results within the scientific community.
To the best of our knowledge, this is the first report about diversity of M. hyopneumoniae in Mendoza province. Results obtained in this work allow drawing a better picture of the genetic diversity of this pathogen in several provinces of Argentina. This is important to understand some epidemiological aspects of EP and to develop strategies to control the disease.
Funding
This study was financially supported in part by PICT 0442/2015 FONCyT-ANPCyT-MinCyT, República Argentina, PPI 2016-2018, 188/2016-UNRC and Dirección Provincial de Ganadería, Ministerio de Economía, Infraestructura y Energía, Gobierno de Mendoza.
Conflict of interest
The authors declare that they have no conflicts of interest.
1. Calsamiglia M, Pijoan C, Trigo A. Applications of a nested-polymerase chain reaction assay to detect Mycoplasma hyopneumoniae from nasal swabs. J Vet Diag Inv. 1999;1:246-51. [ Links ]
2. Charlebois A, Marois-Créhan C, Hélie P, Gagnon CA, Gottschalk M, Archambault M. Genetic diversity of Mycoplasma hyopneumoniae isolates of abattoir pigs. Vet Microbiol. 2014;168:348-56. [ Links ]
3. de Castro LA, Rodrigues Pedroso T, Kuchiishi SS, Ramenzoni M, Kich JD, Zaha A, Henning Vainstein M, Bunselmeyer Ferreira H. Variable number of tandem aminoacid repeats in adhesion-related CDS products in Mycoplasma hyopneumoniae strains. Vet Microbiol. 2006;116:258-69. [ Links ]
4. Dee S, Otake S, Oliveira S, Deen J. Evidence of long distance airborne transport of porcine reproductive and respiratory syndrome virus and Mycoplasma hyopneumoniae. Vet Res. 2009;40:39. [ Links ]
5. Dolso I, Pelliza B, Vissio C, Carranza A, Ambrogi A, Busso J. Lesiones neumónicas halladas en matadero y su asociación con sistemas de crianza de cerdos al aire libre y confinados. In: VI Congreso Nacional de Producción Porcina; 2000. Resumen SP-11, Ciudad Autónoma de Buenos Aires, Argentina.
6. Dos Santos LF, Sreevatsan S, Torremorell M, Moreira MA, Sibila M, Pieters M. Genotype distribution of Mycoplasma hyopneumoniae in swine herds from different geographical regions. Vet Microbiol. 2015;175:374-81. [ Links ]
7. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 1999;41:95-8. [ Links ]
8. Hunter P, Gaston M. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465-6. [ Links ]
9. Mayor D, Zeeh F, Frey J, Kuhnert P. Diversity of Mycoplasma hyopneumoniae in pig farms revealed by direct molecular typing of clinical material. Vet Res. 2007;38:391-8. [ Links ]
10. Nathues H, Grosse Beilage E, Kreienbrock L, Rosengarten R, Spergser J. RAPD and VNTR analyses demonstrate genotypic heterogeneity of Mycoplasma hyopneumoniae isolates from pigs housed in a region with high pig density. Vet Microbiol. 2011;152:338-45. [ Links ]
11. Otake S, Dee S, Corzo C, Oliveira S, Deen J. Long-distance airborne transport of infectious PRRSV and Mycoplasma hyopneumoniae from a swine population infecte with multiple viral variants. Vet Microbiol. 2010;145:198-208. [ Links ]
12. Rebaque F, Camacho P, Parada J, Lucchesi P, Ambrogi A, Tamiozzo P. Persistence of the same genetic type of Mycoplasma hyopneumoniae in a closed herd for at least two years. Rev Argent Microbiol. 2018;50:147-50. [ Links ]
13. Rebaque F, Lucchesi P, Ambrogi A, Tamiozzo P. The use of nested PCR reactions improves genetic typing of Mycoplasma hyopneumoniae from different clinical samples. In Vet. 2016;18: 357-62. [ Links ]
14. Savic B, Ivetic V, Milicevic V, Pavlovic I, Zutic M, Gagrcin M. Genetic diversity of Mycoplasma hyopneumoniae isolates from conventional farrow-to-finish pig farms in Serbia. Acta Vet Hung. 2010;58:297-308. [ Links ]
15. Schlüter PM, Harris SA. Analysis of multilocus fingerprinting data sets containing missing data. Mol Ecol Notes. 2006;6:569-72. [ Links ]
16. Stakenborg T, Vicca J, Butaye P, Maes D, Peeters J, de Kruif A, Haesebrouck F. The diversity of Mycoplasma hyopneumoniae within and between herds using pulsed-field gel electrophoresis. Vet Microbiol. 2005;109:29-36. [ Links ]
17. Stakenborg T, Vicca J, Maes D, Peeters J, de Kruif A, Haesebrouck F, Butaye P. Comparison of molecular techniques for the typing of Mycoplasma hyopneumoniae isolates. J Microbiol Methods. 2006;66:263-75. [ Links ]
18. Tamiozzo P, Lucchesi PMA, Ambrogi A. Mycoplasma hyopneu-moniae genetic diversity in swine farms from Argentina. In Vet. 2011;13:27-35. [ Links ]
19. Tamiozzo P, Lucchesi PMA, Ambrogi A. Monitoring for Mycoplasma hyopneumoniae before and after a partial depopulation program using a typing scheme based on the polyserine repeat motif of p146. J Swine Health Prod. 2013;21:309-12. [ Links ]
20. Tamiozzo P, Zamora R, Lucchesi PM, Estanguet A, Parada J, Carranza A, Camacho P, Ambrogi A. MLVA typing of Mycoplasma hyopneumoniae bacterins and field strains. Vet Rec Open. 2015;2:e000117. [ Links ]
21. Thacker E, Minion C. Mycoplasmosis. In: Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Steneson GW, editors. Diseases of swine. 10th ed. Ames, Iowa: Wiley-Blackwell Publishing; 2012. p. 779 -97. [ Links ]
22. Vranckx K, Maes D, Calus D, Villarreal I, Pasmans F, Haesebrouck F. Multiple-locus variable-number tandem-repeat analysis is a suitable tool for differentiation of Mycoplasma hyopneumoniae strains without cultivation. J Clin Microbiol. 2011;49: 2020 -3. [ Links ]
23. Vranckx K, Maes D, Sacristán Rdel P, Pasmans F, Haesebrouck F. A longitudinal study of the diversity and dynamics of Mycoplasma hyopneumoniae infections in pig herds. Vet Microbiol. 2012;156:315-21. [ Links ]














