Introduction
Porcine cytomegalovirus(PCMV) is an enveloped DNA virus that affects domesticSus scrofapopulations worldwide and constitutes a recognized threat to the swine industry16,19. This pathogen, formally namedSuid betaherpesvirus 2(SuBHV2), is a member of the familyHerpesviridae10. It belongs to the subfamilyBetaherpesvirinaeand has been recently assigned to the genusRoseolovirus8,10. PCMV infection was originally referred to as “inclusion body rhinitis” due to histopathological changes observed in diseased pigs. Viral transmission can occur through the oronasal or transplacental route, with a clinical outcome that strongly depends on the age of the newly infected individuals. Adult animals typically show mild or no manifestations, while respiratory difficulties followed by a severe systemic disease can be developed by neonates and young individuals in naïve herds. Congenital infection can cause reproductive losses owing to fetal mortality16,19. An important characteristic of this agent is its ability to establish latency in those animals that survive primary infection20. It should be noted that PCMV is a major immunosuppressive virus15, thus the disease is likely to be expressed also as a result of coinfections. In that sense, it has been proposed that PCMV could lead to more severe cases of porcine respiratory disease complex (PRDC) by exacerbating the effect of other viral and/or bacterial pathogens9. Similarly, it has been reported that PCMV infection might contribute to the presentation of unusual cases of clinical cystoisosporosis3. No vaccine or specific treatment for PCMV is available19. Despite the fact that free-living swine are known reservoirs and dispersal agents of diverse infectious diseases relevant to pig livestock5,17,18, the PCMV prevalence in wild boars (usually mentioned as wild swine) has been scarcely studied11,21. The aim of this work was to evaluate the PCMV status of the wild boar population that inhabits Northeastern Patagonia, Argentina. The extent to which PCMV was able to spread in our region and also the factors that could influence its epidemiology in natural conditions were also analyzed.
Materials and methods
Wild boar population studied and sampling
The study was conducted on free-living wild boars from Northeastern Patagonia (Buenos Aires and Río Negro provinces), Argentina. Animals were captured by authorized hunters during hunting seasons (Law 5786, decree 2578-1403/05, for Buenos Aires province; Law 2056, decree 633/86, for Río Negro province) between March 2016 and May 2019 (cross-sectional observational study with convenience sampling). Most of the covered areas are part of private fields, where the native vegetation alternates with semi-intensive livestock (bovine, ovine and porcine) and farming production. Information associated with each specimen in terms of geographical point of origin, date of hunting, sex, size and weight was recorded. Wild boars were classified as piglets (less than 6 months of age), juveniles (6-12 months of age) and subadults or adults (more than 12 months of age) according to body size and estimated weight23. No data were available for 19 animals, with regard to precise hunting location, sex and/or approximate age. Data are summarized in Table 1.
Sample with sequenced PCR product: WB02 (sequence A132), WB25 (sequence A123), WB34 (sequence A154), WB56 (sequence A155), WB52 (sequence A113). SeeFig. 2. na: no available data.
Tissues and DNA extraction
Swine tonsils have been reported as appropriate material for molecular detection of PCMV4. These organs were separately recovered from wild boars using clean and/or disposable elements and then stored at −20°C until further processing. After its thawing and mechanical disaggregation, 25-30mg of tissue from each specimen were subjected to total DNA extraction by using the “DNeasy Blood & Tissue Kit” (Qiagen) according to the manufacturer's instructions (proteinase K treatment was carried out for 18h to ensure complete digestion). Purified DNA was kept at −20°C.
Amplification ofSus scrofaand PCMV genomic segments, nucleotide sequencing and phylogenetic analysis
A standard PCR assay to amplify a stretch of theSus scrofabeta-actin gene (ACTB) was optimized according to Deng et al.6. The primers used were F: 5′-CACTTAGCCGTGTTCCTTGCA-3′ and R: 5′-GCGACGTAGCACAGCTTCTC-3′. A 394bp-product was expected according toSus scrofareference sequence NC_010445.4; however the one obtained from wild boar samples was between 400bp and 500bp. PCMV detection was conducted through a nested PCR assay described by Liu et al.14, which amplifies a region of the conservedDPOLgene. In the first round, the primers used were F1: 5′-CGTGGGTTACTATGCTTCTC-3′ and R1: 5′-CTTTCTAACGAGTTCTACGC-3′. In the second round, the primers used were F2: 5′-TGGCTCAGGAAGAGAAAGGAAGTG-3′ and R2: 5′-GACGAGAGGACATTGTTGATAAAG-3′. The expected size of the fragments (according to PCMV reference sequence NC_022233.1) was 769bp and 236bp, respectively. The PCR products were electrophoresed on a 2% agarose gel, stained with GelRed (Biotium), and visualized under UV light. To evaluate the specificity of the viral detection assay, the amplicons of 5 positive reactions were randomly selected (see Table 1) and submitted for direct DNA sequencing (Macrogen, South Korea). The obtained nucleotide sequences were initially compared with those in GenBank and RefSeq databases by using the BLAST program (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The MEGA5.2 software (https://www.megasoftware.net/) package was then used for sequence alignment (ClustalW) and phylogenetic tree construction (Neighbor-Joining method, Kimura 2-parameter model).
Geographical analysis
A map was created with R packages ggmap and ggplot2 using geolocalizations12,22. Geographical distances (km) were calculated from coordinates through the formula: 6371*ACOS(COS(RADIANS(90−Latitude 1))*COS(RADIANS(90−Latitude 2))+SIN(RADIANS(90−Longitude 1))*SIN(RADIANS(90−Longitude 2))*COS(RADIANS(Longitude 1−Longitude 2))).
Statistical analysis
The statistical analysis was conducted by using the Statistix v7.0 program (https://www.statistix.com/). Differences in the distribution of geographic distances were evaluated with the Wilcoxon rank sum test. Comparisons for sex, age, and seasonal variation in PCMV detection rate were made using the Fisher's exact test. An analysis to discard different confounding variables was performed with the Spearman's correlation test.
Nucleotide sequence accession numbers
Sequences determined in this work were submitted to GenBank and were assigned to the following accession numbers: MN831364-MN831368.
Results
Determination of PCMV status in wild boars
An outline of the procedure is depicted inFigure 1a. Tonsils from 65 wild boar specimens were collected, stored, and processed separately to avoid cross contamination. Tissue samples were subjected to total DNA extraction and the recovered genetic material was then used as template in a conventional PCR aimed to amplify a short segment of theSus scrofagenome (Fig. 1b). This assay was carried out as an internal control to evaluate the quality of each DNA sample in order to rule out the presence of enzyme inhibitors and/or excessive DNA fragmentation, which could lead to false negative results in subsequent determinations. In this way, 5% of samples were discarded while the remaining 62 cases were considered for the viral detection stage. A nested PCR designed to render a final product of 236bp in the presence of the PCMV genome was used to assess the status of each individual (Fig. 1c). The analysis revealed that 56% of the wild boar specimens were positive for PCMV, whereas 44% showed no evidence of infection by a criterion restricted to our molecular diagnostic approach (hereinafter referred to as PCMV(+) and PCMV(−), respectively).

Figure 1: Procedure for determining the PCMV status in wild boars. (a) Detail of the steps followed to assign PCMV status to each analyzed animal. (b) Amplification products of the internal control assay. (c) Amplification products of the viral detection assay. Lanes 1-6: different samples analyzed; L: 100bp DNA ladder; NTC: no template control; (+)C: positive control.
Confirmation of virus identity
To verify the specificity of the viral detection assay, the PCR products amplified from 5 different samples were submitted for direct sequencing. As expected, these DNA fragments exhibited high similarity (from 99.49% to 100.00%) at the nucleotide level with the DNA polymerase (DPOL) gene of the PCMV reference strain BJ09 (RefSeq accession no. NC_022233.1, positions 45650-45823). A phylogenetic tree allowed us to visualize that the obtained sequences were intermingled among those retrieved from the database and belonging to previously characterized PCMV strains or isolates (Fig. 2). All these PCMV sequences were placed together in a highly supported branch, which formed a well-defined cluster that was clearly distinct from others that included representative members of the various genera in the subfamilyBetaherpesvirinae.

Figure 2: Phylogenetic position of the viral sequences obtained in this work.DPOLgene sequences from representative members of the subfamilyBetaherpesvirinae(generaRoseolovirus,Cytomegalovirus,MuromegalovirusandProboscivirus) were retrieved from GenBank and RefSeq. PCMV (Suid betaherpesvirus 2/porcine cytomegalovirus): isolate FJ01 (MG696113.1), isolate HN0601 (HQ686081.1), strain B6 (AF268039.2), strain BJ09 (NC_022233.1), strain OF-1 (AF268041.2), strain SC (HQ113116.1). Novel sequences generated in this study: A113 (MN831364), A123 (MN831365), A132 (MN831366), A154 (MN831367), A155 (MN831368). ElBHV1 (Elephantid betaherpesvirus 1, NC_020474.2); HCMV (Human betaherpesvirus 5/human cytomegalovirus, NC_006273.2); HuBHV6A (Human betaherpesvirus 6A, NC_001664.4); HuBHV6B (Human betaherpesvirus 6B, NC_000898.1); HuBHV7 (Human betaherpesvirus 7, NC_001716.2); MuBHV8 (Murid betaherpesvirus 8, NC_019559.1). The evolutionary tree was constructed using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown above the branches (values>50%). The evolutionary distances were calculated using the Kimura 2-parameter model. There was a total of 173 positions in the final dataset.
Geographical distribution of sampling sites and PCMV status
Samples were collected in Northeastern Patagonia, covering an area from 39.0° to 41.1° South Latitude and from 62.4° to 65.9° West Longitude (Fig. 3a). The studied specimens were obtained from 36 different geographical points. These locations are indicated inFigure 3b, which also shows the PCMV status of the specimens collected from each spot. The average distance among all the samples was 103.4±82.8km (mean±standard deviation), 100.3±79.2km for PCMV(−) cases and 107.1±85.7km for those PCMV(+). The statistical analysis revealed no significant differences in the spatial dispersion of the PCMV(−) individuals compared to the PCMV(+) group (Fig. 3c).
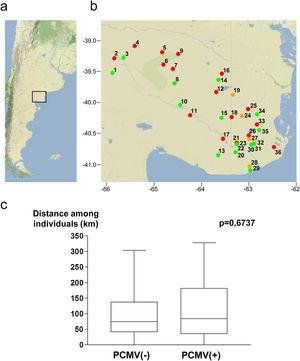
Figure 3: Geographical points of sample collection and PCMV status. (a) Map of Argentina showing the region in which the study was conducted. (b) Detail of sampling locations with information about the PCMV status of the wild boars captured in each site (green spot: only PCMV(−) cases, red spot: only PCMV(+) cases, yellow spot: both PCMV(−) and PCMV(+) cases). Geographical information available for 55 animals. (c) Box and whisker plot depicting the distribution of distances within the PCMV(−) group (n=25) and within the PCMV(+) group (n=30), with medians of 74.5km and 84.0km respectively. A statistical comparison between both groups was performed, the Wilcoxon rank sum test,p-value is shown.
Factors that could affect the PCMV detection rate
Demographic variables such as sex and age were analyzed in relation to PCMV status. It was found that the proportion of infected individuals was very similar between sexes, accounting for 52.4% in females and 53.6% in males (Fig. 4a). However, when divided into age groups, a marked difference was observed between specimens less than 6 months old and all those above this age. Thus, piglets exhibited 88.9% of PCMV(+) cases, while this value accounted for 46.2% in a single category that included juveniles, subadults and adults (Fig. 4b). In order to evaluate variations along an annual cycle, the samples were then separated according to the month of their collection. An increased viral detection rate was found for the period spanning from April to August (Fig. 5a). In contrast, the minimum rate for PCMV occurrence was observed during the first quarter of the year (i.e., January to March, mainly summer), with statistically significant differences regarding the two following trimesters (mainly autumn and winter) (Fig. 5b). To rule out possible confounders in the previous analyses, the correlations between the viral detection rate and (i) the total number of sampled animals, (ii) the number of piglets or (iii) the proportion of piglets were evaluated on a month-by-month basis (not shown). No statistically significant associations were detected (p>0.05, Spearman's correlation test).
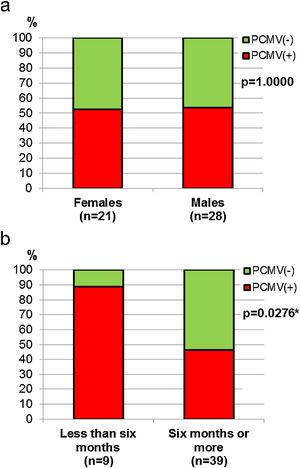
Figure 4: Sex and age in relation to PCMV detection. (a) Comparison of PCMV detection rate between female and male animals. (b) Comparison of PCMV detection rate between age classes. Animals were grouped into two categories: “Less than six months of age” (piglets,n=9) and “Six months of age or older” (juveniles,n=2, and subadults/adults,n=37). Fisher's exact test,p-values are shown (the asterisk indicates a significant difference).
Discussion
Circulation of PCMV in Northeastern Patagonia was assessed through the detection of viral genomes in tonsil tissue from hunted wild boars, which were sampled over an extensive area in Buenos Aires and Río Negro provinces. It was found that 56% of the studied individuals carried the virus, an overall rate that is in line with what was reported for herds of domestic pigs in Asia, Europe, North America and South America7,13,14,19. It should be noted that the prevalence of PCMV is usually estimated using live swine oronasal swabs as samples, a difference with our study that must be taken into account when making comparisons. With regard to the geographical distribution of PCMV-infected animals, the performed analyses showed no evidence of a spatial aggregation of positive cases, suggesting a regular pattern of PCMV spread throughout the entire region analyzed, without major deviations in viral prevalence for any locality. In addition, while no differences between sexes were observed in relation to PCMV status, piglets exhibited a percentage of PCMV positive cases that almost doubles that calculated for all the other growth stages combined. This latter result is similar to previous observations made in domestic pigs, in which the high infection rate of piglets is attributable to the importance of vertical transmission13,14. Finally, it was observed that the frequency of PCMV(+) cases varies along the year, with a sharp increase in autumn. There are references indicating that temperature fluctuations might predispose to PCMV infection and disease development in domestic pigs2. Therefore, in wild boars, the drop in temperatures typical of the autumn weather could modify the PCMV transmission rate and lead to the emergence of newly infected animals, although it is also possible that the main effect of this seasonal factor is to enhance viral replication and particle shedding to simply make PCMV more easily detectable in formerly infected animals.
Taken together, the findings summarized above point to the importance of free-living wild boars as dispersal agents for PCMV. There is no previous information on the prevalence of this virus in domestic pigs within the area covered by this study, where a number of small producers (including families that raise animals for their own consumption) develop semi-intensive livestock practices1. Low-resource conditions generally allow contact between farm pigs and wild boars and, in this context, a shared viral circulation mediated by multiple cross-transmission events can be expected. Further research is required to determine the extent to which this epidemiological dynamic in Northeastern Patagonia can affect both pig production and ecological aspects of wild boar populations.
Funding
This study was financially supported in part by Universidad Nacional de Río Negro (UNRN) [PI 40-C-666, 2017] and Universidad Nacional de Río Negro (UNRN) [PI 40-C-717, 2018].
Conflict of interest
The authors declare that they have no conflicts of interest.












 uBio
uBio 


