The genus Azospirillum is a member of a phylogenetic subgroup in the a-subclass of Proteobacteria. It comprises free-living, diazotrophic bacteria capable of colonizing the internal and external tissues of plant roots2. Azospirillum is one of the most studied genera within plant growth-promoting rhizobacteria - or PGPR - due to its ability to improve the growth and grain yield of many agronomically important crops. Azospirillum spp. are distributed world-wide and have been isolated from the root surface and rhizosphere of various plants, including cereal crops, forage grasses, and cacti13.
Okon and Labandera González10 summarized the results of several field experiments performed in different countries for 20 years and reported that Azospirillum inoculation positively affected plant growth in 60-70% of the cases ana-lyzed, with grain yield increases in the range of 5-30%. Additionally, Díaz-Zorita et al.7 reviewed more than 40 arti-cles, which evidenced 347 cases of positive response to Azospirillum inoculation, with yield increases of 14%, 9.5%, and 6.6% in winter cereals, summer cereals, and legumes, respectively. One of the main mechanisms that explain plant growth promotion by Azosprillum is its ability to produce or metabolize compounds such as phytoregulators10. These compounds include auxins, especially indole 3-acetic acid, gibberellins, cytokinins, nitric oxide, ethylene, and other molecules regulating plant growth under abiotic stress con-ditions such as abscisic acid and cadaverine diamine5.
Azospirillum brasilense Az39 strain was isolated in Argentina in the 1980s within the context of a program car-ried out to identify microorganisms with potential to be used as agricultural bioinputs. This strain is deposited at the PGPR Culture Collection, Instituto de Microbiología y Zoología Agrícola, Instituto Nacional de Tecnología Agropecuaria (INTA), Castelar (Buenos Aires, Argentina). Due to its ability to increase wheat and corn yields, INTA and the Servicio Nacional de Sanidad Agropecuaria (SENASA) recommend A. brasilense Az39 to produce inoculant formulations for corn, wheat, and other non-legume crops. For this reason, this strain is used to manufacture 75% of the inoculants formu-lated in Argentina1,6.
There is an increasing demand for bioinputs to max-imize crop production, and, thus, the quality of these products must be guaranteed. In this context, several sci-entific researchers from public and private organizations of Argentina and abroad established the REDCAI network (Red de Control de Calidad de Inoculantes), a workgroup of the División Microbiología Agrícola y Ambiental (DIMAyA), and member of the Asociación Argentina de Microbiología (AAM). In 2013, the REDCAI network elaborated a hand-book of microbiological procedures for inoculant evaluation3 to promote and standardize the commercial use of inocu-lants. The REDCAI network has already published a complete consensus protocol describing a methodology for detect-ing contaminating microorganisms in inoculant samples and enumerating viable Azospirillum cells by the spread plate technique on a solid culture medium3,4. These two impor-tant parameters for inoculant quality assessment have been evaluated and validated by more than 20 laboratories from Argentina and abroad, belonging to the REDCAI network. However, the routine analysis of many inoculant samples by the spread plate technique described in the REDCAI consensus protocol is laborious and expensive. In this sense, another method for the enumeration of viable cells using fewer Petri plates, with the possibility of including a high number of replicates, has been previously described. This technique, known as the drop plate method9, was proposed for assessing A. brasilense-based inoculants11. The aims of this work were: (1) to standardize and validate the drop plate method for A. brasilense-containing inoculants; (2) to compare the results obtained through this method with those obtained through the spread plate technique previously described in the REDCAI consensus protocol.
To compare the results obtained through the drop plate method and the spread plate technique, both enumera-tion methods were described in detail in a single protocol, which had to be carefully followed across a total of three independent trials, performed in different years using different inoculant samples processed by different laboratories belonging to the REDCAI (Table S1). Standard samples of A. brasilense-based inoculants without trademark identifica-tion were analyzed. The same inoculant sample was further divided and sent to each laboratory involved in each trial. A total of 14 laboratories belonging to the REDCAI participated in the first trial (hereinafter, 'trial 1’). Of them, 7 were private laboratories, and the remaining 7 were laborato-ries belonging to public institutions, including INIA-Uruguay, INTA-Argentina, and several Argentine national universities. The second trial (hereinafter, 'trial 2’) involved 13 private laboratories and 12 laboratories belonging to public institu-tions (INTA, CONICET, and Argentine national universities) comprising 25 operators. This trial constituted an INTERLAB trial, characterized by the inclusion of both trained and inexperienced operators. A total of 17 REDCAI laboratories belonging to the REDCAI participated in the last trial (hereinafter, 'trial 3’), including 4 private laboratories and 13 public laboratories (INTA, SENASA, and Argentine national universities) (Table S1).
The protocol used in all trials included a complete description of the methodology for sample conserva-tion, homogenates and dilution preparation, culture media formulation, incubation conditions, and detection of contaminating microorganisms, according to the REDCAI consensus protocol3. Each laboratory received the inocu-lant sample corresponding to the trial and divided it into three subsamples, which were considered technical repli-cates. To enumerate Azospirillum viable cells through the spread plate technique, 100^l of the dilutions 10-5, 10-6, and 10-7 were sown in duplicate (spreading replicates) in Petri plates containing RC culture medium12, according to the methodology previously described3.
Azospirillum enumeration by the drop plate method in trial 1 was performed using a 10 ^l-drop volume of the dilutions 10-5, 10-6, and 10-7, and a 20^l-drop volume of the dilutions 10-4, 10-5, 10-6, and 10-7, applied by quadrupli-cate (spreading replicates) on Petri plates previously filled with RC culture medium12 (Fig. 1a and b). In trials 2 and 3, bacterial cell enumeration was performed using 20 ^l- and 10 ^l-drop volumes, respectively, of the dilutions 10-4,10-5, 10-6, and 10-7, applied by quadruplicate (Fig. 1a and b). For this enumeration method, water condensation on the agar surface is undesirable because it affects drop absorption and may cause the confluence of different drops. For this reason, the absence of condensed water on the culture medium was carefully checked before the drop distribution. After inoculum application, Petri plates were kept non-inverted until the drops were completely absorbed.

Figure 1: Congo red (RC) Petri plates with 20 pl drop volume of 10-4, 10-5, 10-6, 10-7 dilutions, in quadruplicate. a. Drops distribution on the RC medium surface. b. Typical colonies of Azospirillum spp. in numbers in accordance with the ten-fold dilutions. Photo credits: Dr. Mariana Puente (a) and Dr. Luciana Di Salvo (b).
Petri plates corresponding to both cell enumeration methods were incubated at 28-30 °C for 4 days. Colony counting was performed after the incubation period and repeated 2 days later (6 days after sowing) in order to confirm the typical A. brasilense morphology in the RC culture medium12 (Fig. 1b). Total colony-forming units per milliliter of inoculant (CFU/ml) was calculated taking into account those dilutions which exhibited a range of 5-50 colonies per drop, in the case of the drop plate method, and of 30-300 colonies per plate, in the case of the spread plate technique. For each trial, all participants reported a total of 6 results, 3 for each cell enumeration technique, corresponding to the three subsamples processed as techni-cal replicates. Statistical analyses were carried out using the INFOSTAT/Professional 1.1 software8 to evaluate the reproducibility of these techniques across laboratories and compare both enumeration techniques.
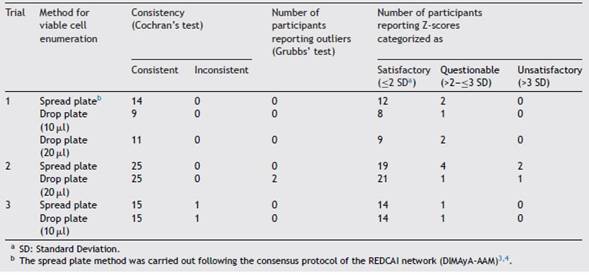
First, the Cochran’s test (0.05%) was performed to determine the consistency of the results reported by each participant of the different trials. This analysis compares the variance of a data set with the total variance of the data sup-plied by all trial participants. Results reported in trial 1 and trial 2 were consistent, while one result reported by one par-ticipant of trial 3 was inconsistent (Table 1). This participant reported highly variable results for viable cell enumeration by the drop plate method, exceeding Cochran’s test critical value. In addition, this laboratory reported highly variable results for viable cell enumeration by the spread plate tech-nique compared with those from the other laboratories. For this reason, all results reported by this laboratory were excluded in further analyses.
Second, the Grubbs’ test (0.05%) was conducted to identify outlier occurrence. This analysis determines if an average value reported by a given laboratory differs signi-ficantly from the average values reported by the remaining laboratories involved in the trial. Only in trial 2, two labo-ratories reported results obtained by the drop plate method that were considered outliers (Table 1). For this reason, the results provided by these laboratories were excluded from further analyses.
Third, data normality was tested by the Shapiro-Wilk’s test. In the case of normal distribution, the Z-score of each trial participant was calculated considering the mean values and standard deviations (SD). There was normal distribu-tion in trials 2 and 3. Conversely, when data distribution was other than normal, Z-scores were calculated considering the median values and the median absolute deviations (MADe). This occurred in trial 1. In the three trials, the Z-scores showed that averages for viable cell numbers were mostly satisfactory (Table 1). In other words, most of the averages were within 2 SD. Only two participants in trial 2 reported unsatisfactory viable cell enumeration averages, one for both enumeration methods and the other only for the drop plate method. These two participants reported viable cell enumeration averages higher than 3 SD. Moreover, in the three trials, some participants reported ''questionable’’ viable cell enumeration averages (Table 1) by the spread plate technique (two participants in trial 1, four participants in trial 2, and one participant in trial 3), by the drop plate method usinga 10 pl-drop volume (one participant in trial 1 and one participant in trial 3), and by the drop plate method using a 20 pl-drop volume (two members in trial 1 and one memberin trial 2). ''Questionable’’ viable cell enumeration averages were within 2 and 3 SD. Unsatisfactory and questionable viable cell enumeration averages were excluded from the comparative analyses between both enumeration methods in all cases. Thus, only the results obtained by those participants who reported consistent, robust, and satisfac-tory values by both enumeration methods in the three trials were included.
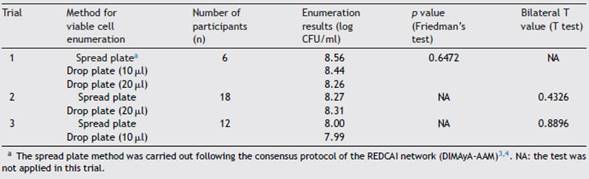
Considering that data from trial 2 and trial 3 had a normal distribution, but those from trial 1 did not, the comparison between both enumeration methods was performed by the paired-sample T-test and Friedman’s test, respectively.Table 2 shows the number of participants whose results were included in the comparison. In addition, the average values for viable cell numbers found in different samples of Azospirillum inoculants, evaluated in the three trials by both enumeration methods, are shown. No differences between the drop plate method and the spread plate technique were observed (Table 2). Based on these results, we concluded that Azospirillum viable cell enumeration performed by the drop plate method using 10 pl- or 20 pl-drop volumes is equivalent to the Azospirillum viable cell enumeration performed by the spread plate technique, previously validated and published3,4.
The drop plate method validation performed in this work was achieved thanks to the contribution of many REDCAI members from different institutions and geographical areas of Argentina and abroad. This method allows to include more replicates using fewer Petri plates and culture medium amounts, making it a more cost-effective method than the spread plate technique9. Furthermore, fewer Petri plates and culture media consumption implies a reduction in waste amounts, which would result in reduced environmental impact. With regard to the complexity of the enumeration techniques, this work shows that the only requirement to carry out the drop plate method is to have minimal training in microbiological techniques. These trials, especially trial 2, involved both trained and inexperienced participants, and even so, reliable results were obtained. Based on this work, we can say that the inexperience in the drop plate method is not expected to constitute a limitation if a clear and com-prehensive protocol is available to facilitate the work and to guarantee reliable results, with the commitment to comply with this consensus protocol.
Finally, it is important to point out that the performance and participation of individual laboratories were confidential. Additionally, commercial inoculants fraction-ated in white bladders without trademark identification were distributed because the aim of this work was not to evaluate the quality of the inoculants. This work is in line with the goals of the REDCAI network, among which, the standardization and validation of consensus protocols for the quality assessment of microbial inoculants stand out. The validation of the drop plate method for other Azospirillum species remains to be accomplished. We validated the drop plate method as a viable cell enumeration technique for A. brasilense. Therefore, this method will be included as an alternative method for the quality control of Azospirillum-containing inoculants in the consensus pro-tocol of the REDCAI network (DIMAyA-AAM).
Conflict of interest
The authors declare that they have no conflicts of interest.
Acknowledgements
We are grateful to the former coordinators of the Azospiril-lum workgroup in the REDCAI, Dr. Rosana Massa, Dr. Cecilia Creus and Dr. Fabricio Cassán, to REDCAI coordinator Dr. Silvia Toresani, and to the companies which provided us the inoculant samples used in our trials. We thank Luisina Andriolo, Lucas Dalmasso, Andrés Laurent, Romina Molina, Gisel Peralta, María Eugenia Schiavon, and Juan Silberman for their contributions to this work. We specially thank Ing. Agr. Enrique Rodríguez Cáceres, whose generosity in sharing his large experience working with Azospirillum genus has been valuable enriched our workgroup discussions. Finally, we are also grateful to editors and anonymous reviewers for their comments and suggestions.












 uBio
uBio