Contagious Ecthyma (CE) is a severe exanthematous dermatitis affecting small domestic ruminants such as sheep and goats, although it can also be detected in wild animals6. CE is caused by the Orf virus (ORFV), member of the Poxviri-dae Family, genus Parapoxvirus (PPV)3. ORFV has a linear double-stranded DNA genome, of approximately 135 kbp in size and exhibits an unusual high GC content (~64%). The central region of the genome contains 88 genes which are present in the subfamily Chordopoxvirinae and mostly occur in a common order and orientation. The terminal regions of the genome are variable and there are genes associated with virulence, pathogenesis, tropism and immune response modulators (orf001 to orf008 and orf112 to orf134).
ORFV is an epitheliotropic virus that causes lesions on the lips, nostrils, oral mucosa and udders of sheep and goats. ORFV can remain infective for many weeks, especially in scabs, thus enabling widespread infection in the flock10.
CE is also a zoonotic disease that can spread to humans after contact with infected animals or contaminated fomites2. Human skin lesions due to ORFV are usually self-limitingand resolve spontaneously within 4-6 weeks, except in immunocompromised individuals5.
Human ORFV infections are not usually reported in South American clinical practice. Considering the widespread dis-tribution of CE in flocks all over the region1,11,12, ORFV infection in humans may be underdiagnosed, which may lead to overtreatment or unnecessary medical procedures15.
The first human ORFV case diagnosed in Argentina was detected in a 52-year-old female sheep keeper from a farm in Villarino Department (south of Buenos Aires province). The flock of the farm had suffered a CE outbreak (lamb skin samples were identified as sMbu15) that affected more than 90% of young lambs (<3 months old) (Fig. 1A) weeks before the disease was confirmed in the keeper. The patient showed erythematous papules on the back of both hands (Fig. 1B) that evolved into inflammatory nodules with a diameter of approximately 2cm (Fig. 1C). Initially, she attended a pri-mary health care center, where the signs were attributed to a complication of her diabetic condition. Two weeks later, the lesions progressed to the left forearm, with erythe-matous, edematous linear appearance and evident pain, compatible with lymphangitis. Once ORFV infection was con-firmed in lambs from the flock she cared for, she was advised to return to the health center where a skin sample was biop-sied (hMbu15) and sent for analysis. Informed consent was obtained from the individual that participated in the study.
In Chile, the first human ORFV case was reported by Flores et al.4 Briefly, a young veterinary student received a goring wound in her right hand when she was performing a clinical examination of apparently healthy goats from a flock. Two weeks later, she noted the appearance of typical EC lesions and consulted a dermatologist, who biopsied the skin samples (hChi17) for PCR diagnosis. No further analysis was performed.
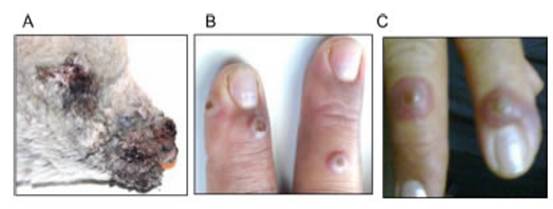
Figure 1: Lesions caused by ORFV infection in an Argentine rural worker. (A) Photograph of a dramatic ORFV infection in a 3-month-old affected lamb. (B) Three erythematous papules are observed on the back of the right hand. (C) Few days later, papules evolved into inflammatory nodules measuring 2cm in diameter.
In this work, we analyzed hMbu15 and hChi17 and compared them with other ruminant ORFV strains by using different genes: orf011, orf020, orf109, and orf127. We present the first human CE case confirmed in Argentina along with a phylogenetic analysis of its etiological agent.
First, we confirmed an ORFV infection in a human scab sample (hMbu15) by using an orf045 internal region gene amplification by PCR. This diagnostic PCR proved to have 100% efficiency in detected viral genomes in dif-ferent samples8. Subsequently, DNA samples hChi17 and hMbu15 were used as a template for PCR reactions to amplify an orf011 gene fragment and the complete open reading frame of orf020, orf109 and orf127 genes, as previously described12. The orf011 gene encodes a highly immunogenic major envelope protein B2L. The internal region of the orf011gene (nt 388 to 981) has been mostly used for the phylogenetic analysis because it is the virus sequence most deposited in databases, thus allowing strains from all over the world to be compared. Furthermore, we analyzed the orf020 and orf127 genes, which codify virulence factors VIR and vIL10 respec-tively, and orf109 because it encodes an envelope mature protein (EEV), one of the most variable genes within the PPV genus. The sequences were submitted to the GenBank databases under the following accession num-bers: orf011internal region: MH161454, MH161455; orf020: MH161456, MH161457; orf109: MH161458, MH161459 and orf127: MH161460, MH161461.
The sequence of the orf011 internal region gene7 is com-monly used for the OFRV phylogenetic analysis9. The identity matrix showed that hMbu15 was 100% identical to sMbu15, while hChi17 showed 99.8% and 100% identity with other ORFV strains isolated from goats of Rio Negro (gCom14) and San Luis (gCnc15) provinces in Argentina, respectively.
As expected, a maximum likelihood (ML) phylogenetic tree analysis resulted in a group that contained hChi17 together with the above mentioned ORFV isolates from Argentina (gCom14and gCnc15) and Brazil (NE2, NE1, Bahia and SJ1) (Fig. 2A). This group contains exclusively goat ORFV strains, with a significant bootstrap support (Fig. 2A). On the other hand, hMbu15 constitutes a heterogeneous group together with other ORFV isolates obtained from sheep (sMbu15, sBal15, sChu14, sPil14, sPi13) and goats (gNeu14) from Argentina as well as isolates from goats in Brazil (MT05, 578/08, 561/11 and 27/12), Uruguay (UY19/10) and North America (orf-mu, orf-ta and orf-sh) (Fig. 2A). This particular tree group exhibits many branches with low or interme-diate bootstrap values, probably due to a high nucleotide sequence identity among all included samples in the phylo-genetic analysis13,14.
To improve this analysis, we extended the study to other ORFV genes: orf020 and orf127, which were described as virulence factors, and orf109, which codifies an extracel-lular mature virion protein that is orthologous to vaccinia virus A33R protein3. In accordance with the finding using the orf011 gene, hMbu15 and sMbu15 were 100% identical in A. Peralta, C. Flores-Olivares, A. Verna et al. all genes analyzed, which confirms the origin of the human infection. Moreover, the analysis also revealed 100% iden-tity between hChi17 and the Argentine gCom14 and gCnc15 in orf109 and orf127. However, the orf020 gene exhibited some variability between hChi17 with gCom14 and gCnc15 (0.2 and 0.5% respectively). Subsequently, a ML phylogenetic tree was constructed with concatenated orf011, orf020, orf109 and orf127 sequences to simplify the visualization of the global analysis (Fig. 2B).

Figure 2: Phylogenetic analysis of South American human ORFV strains. The phylogenetic relationships were constructed by the maximum likelihood algorithm using the MEGA 6.0 software. All positions with less than 50% site coverage were eliminated. The tree with the highest log likelihood is shown. Initial trees for the heuristic search were obtained automati-cally by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances. Numbers at nodes represent% of 500 bootstrap replicates. White circles indicate Argentinian sheep and goat samples, the dark circle indicates the Argentinian human sample, while dark triangle, the Chilean human sample. (A) Phylogenetic analysis based on 517 nucleotide sequence positions of the orf011 internal region; theTamura-3 parameter was selected as the best-fit evolution model. (B) Phylogenetic analysis based on 2046 nucleotide sequence positions of the orf011, orf020, orf109and orf127concatenated sequences; the Kimura-2 parameter was selected as the best-fit evolution model. Only strains with the four molecular marker sequences available at the Gene Bank were included.
As expected, hMbu15 and sMbu15 were identical and formed a branch with other ORFV strains from Buenos Aires province (sBal15 and sCh97), with 100% bootstrap support, and separated from other strains detected in Argentina (Fig. 2B). This finding could indicate the heterogeneity of ORFV strains from different regions in Argentina.
The phylogenetic analysis with four molecular markers along the genome also confirmed no close phylogenetic relationship between hMbu15 and hChi17 samples. Surpris-ingly, hChi17 was closely related to other goat ORFV strains from Río Negro (gCom14) and San Luis (gCnc15) provinces in Argentina. A phylogenetic analysis obtained from the four concatenated molecular markers showed that gCom14, gCnc15 and hChi17 form a group with high confidence sup-port (Fig. 2B), which indicates a close relationship between the three isolates. Moreover, they were 100% identical in two of the four markers studied (orf109 and orf127 genes), which may indicate that these strains share a common origin. There is a natural border between Argentina and Chile (The Andes range of mountains), but apparently this barrier does not prevent the exchange of animals or contaminated fomites that move the virus from one region to another. Nev-ertheless, studies including more ORFV strains from Chile and Argentina are necessary to confirm this hypothesis and further studies could help to establish new policies on the ruminant movements between both countries.
The first human CE case from Argentina was initially mis-diagnosed by medical practitioners. This is a common finding in the region since healthcare staff are usually unaware of the occurrence of this zoonosis. It is necessary to warn physi-cians about the probability of ORFV infection in humans, especially when they are in contact with susceptible ani-mals. Zoonotic diseases, such as CE, could represent a threat to human health as was widely demonstrated by the COVID-19 pandemic.
This study presents the first molecular comparison of human ORFV strains from Argentina and Chile. The obtained data increase the knowledge of the ORFV strains circulating in the region at a molecular level and show the importance of performing an update about human CE in the region, both for workers with exposure risk and healthcare providers.
Conflict of interest
The authors have no conflict of interest to declare.
Authors' contributions
AP, CFO, and GC contributed to the conception and design of the study. AP performed the PCRs for the four molecular markers and the sequence analysis. GAK per-formed the phylogenetic analysis. AV, EGA and AO performed diagnostic PCR. CFO, EO and CM characterized the outbreaks in the field, identified human cases and tookepidemiological data.
All authors contributed to manuscript revision, read and approved the submitted version.
Acknowledgment
Financial support was provided by PNBIO1131032 and PNSA1115055, Instituto Nacional de Tecnología Agropecuaria (INTA), Argentina.
Received 30 December 2021
accepted 18 July 202












 uBio
uBio 

