Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Latin American applied research
versão impressa ISSN 0327-0793
Lat. Am. appl. res. v.35 n.4 Bahía Blanca out./dez. 2005
Characteristics of a methanogenic biofilm on sand particles in a fluidized bed reactor
M. Mussati1, C. Thompson2, M. Fuentes3, P. Aguirre4 and N. Scenna5
INGAR-Instituto de Desarrollo y Diseño/CONICET, Avellaneda 3657,(3000) Santa Fe, Argentina.
{1 mmussati; 2 cthomps; 3 mfuentes; 4 paguir; 5 nscenna}@ceride.gov.ar
Abstract — The typical microbiological groups reported in literature under the experienced environmental conditions are present in the biofilm structure investigated on sand particles. This is concluded based on microorganism morphology analyzed by scanning electron microscopy. Methanosarcina sp. (acetate consumers) and filamentous microorganisms with morphology similar to Methanospirillum sp. (H2-utilizing archaea) are distinguished among methanogens. Based on acetate levels and microorganism's threshold concentration, a predominance of Methanosarcina mazei rather than Methanosarcina barkeri is concluded. Among acetogens, bacillus morphologically similar to the syntrophic acetogenic Syntrophobacter wolinii are observed together with Desulfovibrio sp. (hydrogen-utilizing, sulfate-reducing). With respect to acidogens, short and long rod-shaped bacteria, diplococcus in chains (probably Streptococcus-like bacteria) and filamentous bacilli are morphologically distinguished but cannot be characterized from this study. A methanogenic biofilm fluidized bed reactor inoculated with the biofilm population investigated showed process efficiencies up to 98% of chemical oxygen demand reduction for treating an acetate-based substrate.
Keywords — Methanogenic Biofilm Composition. SEM. Reactor Performance.
I. INTRODUCTION
Microbiological processes have been extensively applying for the removal of inorganic and organic compounds from wastewaters. In general, the treatment processes have to be stable, low in cost and to provide an effluent quality to comply with the increasingly stringent discharge standards. These needs can be met by either optimizing the operation of existing wastewater treatment plants, which are mainly based on the conventional aerobic processes, or by developing novel bioprocesses.
During last decades the anaerobic digestion has been considered as an attractive biotechnological process for degrading a variety of polluting organic wastes. However, the benefits of this technology are not restricted to only removal of contaminants. Indeed, energy savings can be obtained by computing the biogas produced (fuel methane) in a global process analysis.
The anaerobic degradation is a process carried out by one of the most complex known interactions among microbial populations interacting in a food web (Davey and O'toole, 2000). Briefly, the process consists of three stages: (a) the first one (liquefication) involves the hydrolysis and conversion of insoluble complex material to soluble compounds and the reduction of polymers to monomers; (b) acidogenesis involves the fermentation of the monomers into a variety of end-products, which include volatile acids (acetate, propionate, butyrate, formic acid), alcohols, carbon dioxide and hydrogen; and (c) methanogenesis: the end-products of the fermentation process are then converted by another group of anaerobes into methane and carbon dioxide with trace quantities of other gases (hydrogen sulphide, ammonia, nitrogen, mercaptans and amines) (Ghaly, 1996).
The application of the anaerobic biotechnology required the overcoming of some difficulties at the early development stages, mainly related to the anaerobic digestion process itself, i.e. the slow growth rate of the anaerobic methanogenic microorganisms. This disadvantage discouraged its application to continuous systems operating at short retention times (high dilution rates) since the microorganism washout takes place under such conditions. This drawback was technologically overcome by decoupling the hydraulic retention time from the solid (microorganisms) retention time by development of granular biomass or attaching or immobilizing the microbial consortium on inert support materials (biofilms). In biofilms, the microorganisms are embedded in a matrix of organic extracellular polymeric substances produced by the microbial activity and also may contain inorganic or abiotic substances.
In general, the relative population levels of the microbial groups (acidogens, acetogens, methanogens) and the species of each group present depend on the wastewater characteristics as well as on the operational and environmental conditions, e.g. substrate type and organic loading rate, system configuration, inert support material type and shear stress acting on it. Any stress or disturbance on the system may lead to a change in species types and their relative population levels, which is ultimately reflected in the reactor performance. Then, it becomes evident the importance of a well-balanced and mature biofilm development and methods for monitoring its structure. The scanning electron microscope (SEM) can be used to obtain knowledge of both morphology and composition of the populations of anaerobes coexisting in the biofilm structure of digesters (Lazarova and Manem, 1995; Araujo et al., 1998; Díaz et al., 2003; Montenegro et al., 2003; Yang et al., 2004).
In this paper, the characteristics of a methanogenic biofilm developed in a fluidized bed reactor fed with a milk powder-based substrate are investigated through visual characterization by scanning electron microscopy (SEM). Sand particles are the support material used. Then, the performance of a second bioreactor seeded with an inoculum obtained from the former one and fed with an acetate-based synthetic substrate is reported.
II. MATERIALS AND METHODS
A. Bioreactors
Two lab-scale anaerobic fluidized bed reactors were used to carry out the experiments (hereafter named R1 and R2), which were loaded with sand as support material.
Bioreactor R1. The bioreactor R1 consists of a 0.7 m high acrylic cylinder with 0.0816 m inner diameter. The static bed was 0.16 m high and the fluidized bed height was fixed according to the operation conditions needed, e.g. 0.21 m high during the inoculation stage to have about 30% bed expansion and a reaction volume of 1.1L. The upper zone of the reactor is a gas chamber that accumulates the biogas produced. The liquid volume of the whole system is approximately 4.5 L. The liquid stream is conducted to a cyclonic separation system consisting of a 0.3 m high cylinder with 0.0816 m inner diameter, where solid particles and a fraction of the dissolved gases are separated from the liquid stream. The liquid phase occupies 40% of the total volume and the rest by biogas. The gaseous stream out-coming from the cyclone is accumulated in a pressurized collection system, sealed with an acid solution (pH 2.5). The solid material accumulated at the cyclone bottom can be either recycled to the reaction zone or wasted. Most of the liquid contained in the cyclone is recycled back to the reaction zone, while the remaining fraction is the cleaned stream leaving the system. The liquid level in the cyclone is fixed by the relative position of the effluent discharge point. The fresh feed is injected at the liquid recycle line by a syringe-type pump. The reactor pressure is fixed by over-pressure in the gas collection system.
Bioreactor R2. The bioreactor R2 is a column consisting of a 2.0 m high acrylic cylinder, whose inner diameter is 0.065 m. The separation compartment placed over the column is a 0.180 m high cylinder with a 0.145 m inner diameter, where gas accumulation and particle sedimentation take place. The effluent discharge, the feed input and the recycle suction point are also placed in this compartment. The setup used for bioreactor R2 is schematized in Fig. 1 and the bioreactor specification data are listed in Table 1.
B. Inert support material.
A sample of sand used in bioreactors R1 and R2 was meshed using the Tyler sieve series. The particle size distribution is listed in Table 2. The material specific surface and the surface volume mean diameter are calculated as in McCabe et al. (1993) and Perry and Chilton (1973). These values and other inert support characteristics are included in Table 3.
Table 1. Specification data for bioreactor R2

Table 2. Results of sand sample meshing.

Table 3. Characteristics of the inert support material.

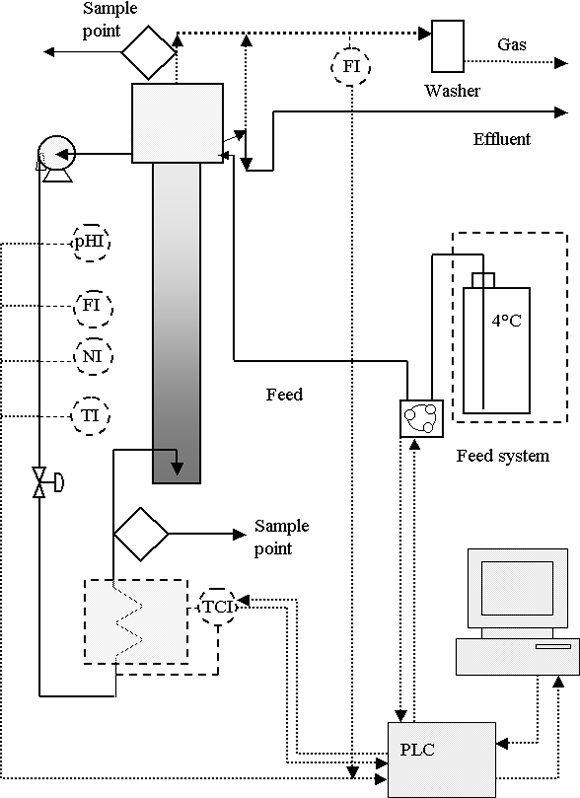
Figure 1. Experimental set-up for bioreactor R2.
D. Analytical Methods
The amount of biogas produced by the bioreactor was measured using a water replacement method. The gas composition was analyzed by a gas chromatograph (Hewlett Packard 6890) equipped with a thermal conductivity detector and a 3 m carbosphere column. Hydrogen was used as carrier gas at 20 mL min-1. The column was operated at 150oC. The injector and detector temperatures were 100 and 230oC, respectively.
The effluent chemical oxygen demand (COD) was measured according to the HACH potassium dichromate method approved by USEPA (Cat. 21259-15, 0-1500 ppm). The concentration of the released Cr3+ ions was determined by spectrophotometry (Metrolab 330). The effluent pH was measured using a digital pHmeter (Horiba D-12). Some samples were also measured according Standard Methods for COD to evaluate the approximation obtained with the HACH kit. The results were satisfactory.
The acetic, propionic and butyric acid (VFA) concentrations were measured by a high pressure liquid chromatograph (HPLC) Hewlett Packard Model Series 1050 equipped with a UV-VIS detector (wavelength: 215 nm). A 20 µL sample volume was injected into a Spherisorb ODS-1 (C18) Classic 5U (250 x 4.6 mm) column (Alltech, Deerfield, IL). The mobile phase consisted of 50% acetonitrile-50% water, sulfuric acid 0,01% (pH 3) at a flow rate of 0.7 ml/min.
E. Scanning Electron Microscopy
Samples for scanning electron microscopy were fixed in 2.5% glutaraldehyde in a phosphate buffer solution (PBS) 0.1M pH 6.8 for 24 hr at 4oC. The fixed samples were washed twice with PBS and dehydrated with increasing ethanol concentrations (25, 50, 70, 80, 90, 95, 100% vol vol-1) for 10 min. each one, using a 0.45μm pore size cellulose nitrate MicroFiltration Systems (MFS) plate filter. The samples were placed on glass cover slides, which were mounted on aluminum stubs. The samples were coated with a gold layer in an argon atmosphere in a vacuum evaporator Veeco, prior to examination with a JSM-35C scanning electron microscope (JEOL, Tokyo, Japan), operated at a 15 kV acceleration voltage.
F. Experimental Protocol
Inoculum and Inoculation Procedure
- Inoculum source. The bioreactor R1 was inoculated with mixed liquor obtained from a 15 L anaerobic continuous stirred tank reactor (ACSTR) fed with a milk powder- and acetate-based synthetic substrate (0.4 and 1.0 g COD L-1, respectively). The system pH was adjusted to 6.5 by adding a sodium bicarbonate solution. The ACSTR reactor was prior inoculated with mixed liquor obtained from different anaerobic sludge sources. The selected substrate composition and the initial pH value are intended to favor the growth of the methanogenic microorganisms. After inoculation, the ACSTR was operated at 10, 7 and 4-day retention times during 40, 28 and 24 days, respectively, at 28±2oC. At day 7 of the latter period, the pH was adjusted to 5.5 by an acidic solution. After 1 week, the pH was raised to 8.0 by a sodium bicarbonate solution. After 10 days, the retention time was set to 10 days and the influent COD was duplicated (0.8 and 2.0 g COD L-1 of milk powder and acetate, respectively). After 1 week, the retention time was decreased to 2 days during 7 days. The last two disturbances were repeated 8 times at different temperatures (between 22 and 34oC) using the initial substrate concentration (0.4 and 1.0 g COD L-1 of milk powder and acetate, respectively). The system capability for producing methane was periodically verified. This operation policy after inoculation is intended to render a microbial consortium capable of resisting the shocks and overloadings often acting on bioreactors in real operation conditions.
- Inoculation of biofilm reactor R1. 50 ml of settled biomass from the ACSTR reactor was used to inoculate R1 when fresh sterilized substrate was recirculating in the system. The synthetic substrate consisted of 1.77 and 1.07 g COD L-1 of milk powder and sodium acetate, respectively, resulting in a pH 6.5. Initially, low retention times were adopted in order to facilitate the fixation of microorganisms with better adhesion properties. Then, the organic loading rate was fixed at 3.34 g COD d-1 L-1 of reactor during 42 days for microbial fixation. The bed expansion was fixed at 30%.
- Inoculation of biofilm reactor R2. Fractions of both solid and liquid phases taken from R1, solid material deposited at the cyclone bottom of R1 (sand particles and pieces of released biofilm), and the out-coming stream of R1 were used as inoculum for reactor R2. The substrate consisted initially of milk powder in order to provide the inoculum with similar environmental conditions as reactor R1. The milk powder-based feed was progressively replaced by acetate until reaching 95% of the total COD. The rest consisted of milk powder in order to supply the micro and macronutrients necessary for microbial growth.
Load policies for the biofilm reactors. Disturbances.
Reactor R1. After inoculation and microbial fixation, the substrate fed to reactor R1 consisted of milk powder with a concentration of 1.44 g L-1. The organic loading rate was 2.8 g COD L-1 d-1. The system pH ranged between 6.5 and 7.5 and the temperature was kept constant at 36±2oC.
Reactor R2. The performance of the reactor R2 to disturbances on the acetic acid concentration and the volumetric feed flow rate was investigated. The experiments were carried out at 36±1oC. The feed to reactor R2 consisted initially of 2.0 g COD L-1 (90% acetate and 10% milk powder plus an amount of sodium bicarbonate for pH adjustment) at 3.6 L d-1. A step-type disturbance on the inlet acetic acid concentration was the first perturbation introduced (P1) increasing the influent COD with 50% (from 2.0 to 3.0 g L-1). The feed flow rate was kept constant at 3.6 L d-1. The disturbance was applied when reactor operated close to steady state conditions corresponding to 2.0 g COD L-1 and 3.6 L d-1. After 28 days under P1 conditions, the inlet COD concentration was increased from 3.0 to 4.0 g L-1 (P2), keeping the same feed flow rate (3.6 L d-1) during 20 days. Finally, disturbance P3 consisted of a step in the feed flow rate, from 3.6 to 7.2 L d-1, keeping the input concentration at 4.0 g COD L-1. The disturbances applied are depicted in Figs. 6, 7 and 8.
III. RESULTS AND DISCUSSION
A. Biofilm Structure
In this subsection, aspects of the biofilm developed using bioreactor R1 are discussed. As mentioned, bioreactor R1 was fed with a milk powder-based substrate, which is constituted approximately by 44.8% of lactose. Methanization of lactose needs a cooperative biological activity from three bacterial types: acidogens, acetogens and methanogens (Yu and Pinder, 1993). Yang and Guo (1991) have found that propionate and acetate were the only two major organic intermediates found in the methanogenic fermentation of lactose. These authors proposed a three-step reaction mechanism. First, lactose is hydrolyzed and fermented to, mainly, propionate, acetate, H2 and CO2 by acidogenic bacteria. In the acetogenic phase, propionate is degraded to acetate, CO2 and H2. In the last phase, these products are used for biogas formation (CO2 and CH4) by methanogens. These results agree with the ones reported by Yu et al. (2002) using full cream powdered milk as substrate. Based on this, following the three microbial groups involved were investigated in the resultant biofilm structure. Since methane production is usually the rate-limiting stage, the focus is first on methanogens.
Methanogens. These microbes belong to the domain archaea (Le Mer and Roger, 2001). Two different microorganisms are found within the methanogenic archaeas that can convert the acetic acid into CH4 and CO2: Methanosarcina sp. and Methanosaeta sp. In fact, these two archaea genera are the only known methanogens capable to metabolize acetate (Speece, 1996) and compete with each other for acetate. Unlike Methanosaeta sp, which only grows on acetate, Methanosarcina sp. is also able to grow on other substrates such as hydrogen, formate and methanol. In addition, Methanosarcina sp. has a much higher maximum specific utilization rate but a lower affinity for acetate (higher Monod saturation constant KS values) than Methanosaeta sp. Based on this, the latter predominates at low acetate concentrations; while Methanosarcina sp. predominates at higher acetate concentrations because of their competitive advantage, if specific trace metal bioavailability is satisfied (Speece, 1996; Janssen, 2003; Batstone et al., 2004). The threshold concentration is the minimum concentration below which a species is no longer able to degrade a particular substrate (Großkopf et al., 1998), and can be used as an approximated indicator of the predominance of species in a system. According to Jetten et al. (1992) Methanosaeta sp. shows a much lower minimum threshold for acetate utilization (7-70 μM) than Methanosarcina sp. (200-1200 μM). Großkopf et al. (1998) reported acetate concentration thresholds of 850 μ M and 310 μ M for the strain Methanosarcina barkeri MST and Methanosarcina sp. strain VeA23, respectively. Westermann et al. (1989) estimated threshold values for Methanosarcina barkeri, Methanosarcina mazei and Methanosaeta sp. of 1110, 400 and 65 μM, respectively. This is consistent with the evidence that Methanosaeta sp. is found in environments with low acetate concentrations (Jetten et al., 1992).
With respect to pH preferences of species, Brummeler et al. (1985) reported that Methanosaeta soehngenii has an optimum pH at 7.8 and no activity below pH 6.8; while Methanosarcina sp. forms methane in a much wider pH range (5 to 8).

Figure 2. (1) Methanosarcina sp.; (2) filamentous microorganisms morphologically similar to Methanospirillum sp.; (3) comma-shaped microorganisms resembling Desulphovibrio sp.; (4) bacillus morphologically similar to Syntrophobacter wolinii.
In the bioreactor here investigated, the measured acetate concentrations ranged between 390 and 925 μM and pH between 5.9 and 6.4. Then, based on these results a predominance of the Methanosarcina sp. rather than Methanosaeta sp. in the composition of the biofilm developed is expected. That is indeed the case as can be observed from micrographs shown in Figs. 2 and 3, where clusters of microorganisms morphologically similar as Methanosarcina sp., large archaeal cocci in groups of four individuals can be distinguished. Based strictly on threshold values (since they are morphologically identical), it could be concluded that M. mazei predominates rather than M. barkeri in the structure of biofilm developed under the conditions experienced. Inside granules, which are other type of microbial aggregates, predominance of Methanosaeta sp. can be found (Brummeler et al., 1985; Batstone et al., 2004) since a concentration profile is established due to both substrate consumption by microorganisms close to the granule surface and mass transfer resistance. Moreover, Methanosaeta sp. plays a critical role during the granule formation (Bhatti et al., 1995). As shown in Fig. 4, that is not the case here investigated since the biofilm developed on sand particles is characterized by a thin thickness and, consequently, the diffusion limitations are not important.

Figure 3. Methanogens: Methanosarcina sp
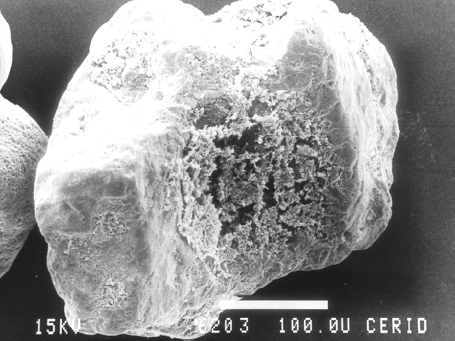
Figure 4. Representative sand particle partially covered with an anaerobic biofilm of thin thickness.
Acetogens. Propionate and butyrate are thought to be converted to acetate only by syntrophic acetogens in concert with hydrogen-utilizing methanogens (Lowe et al., 1993). Syntrophobacter wolinii was the first syntrophic propionate-degrading culture isolated from methanogenic enrichments from an anaerobic municipal sewage digester in association with hydrogen-utilizing bacteria (Lowe et al., 1993). Propionate-oxidizing Syntrophobacter-like bacteria have been identified in microcolonies in intimate association with methanogens (de Bok et al., 2004) such as Methanospirillum sp. (Boone and Xun, 1987), or associated with hydrogen-utilizing, sulfate-reducing Desulfovibrio sp. Syntrophobacter sp. are non-sporulating, Gram-negative, rod-shaped bacteria that occur singly or in pairs with some short chains and filaments. The cell width is 0.6 to 1 µm (sometimes irregular, especially in filaments) and 1.0 to 4.5 µm long (Boone and Bryant, 1980). As mentioned, a methanogenic archaea isolated with Syntrophobacter sp. is Methanospirillum hungatei, which can produce methane from H2 and CO2. They are curved rods measuring between 0.5 and 7.4 µm. Cells most often occur in filaments of 15 to several hundreds µm long. The optimum temperature and pH range between 30-37oC and 6.6-7.4, respectively (Ferry at al., 1974), which are the operation conditions here experienced. In Fig. 2, filamentous bacteria with morphology similar to archaea Methanospirillum sp., very few comma-shaped microorganisms resembling Desulphovibrio sp. and bacillus morphologically similar to Syntrophobacter wolinii can be observed.
Acidogens. Many hydrolyzing microorganisms and acidogens can coexist in anaerobic methanogenic biofilms but very little information is available on the characterization of the bacteria involved in the acidogenic phase (Bramucci and Nagarajan, 2000; Zellner et al., 1999). This is even worst when the characterization is only based on SEM analysis due to the large variety of species with similar morphology. Stafford et al. (1980) and Miyamoto (1997) indicate that bacteria belonging to Clostridium sp. have been isolated from different types of anaerobic digesters but without specifying the effluent type treated. Clostridium sp. are responsible for most of the extracellular lipase and protease produced, and convert the metabolites into acid products. These strict anaerobic microorganisms are rod-shaped, 2.8-3.0 mm long and 0.5-0.6 mm wide. The optimal growth temperature and pH vary between 35-37 oC and 4.5-7.0, respectively, (Zigová et al., 1999). Clostridium sp. has been isolated from a swine manure digester (Iannotti et al., 1982) and C. collagenovorans (optimum pH 6-8) from a sewage digester (Lowe et al., 1993), which are the operation conditions but not the substrate type here investigated.
Different kind of bacteria such as short and long rods, diplococcus in chains (probably Streptococcus-like bacteria) and filamentous bacilli can be morphologically distinguished in Fig. 5 but cannot be characterized from this study.
B. Bioreactor performance
In this subsection, the performance of bioreactor R2 is analyzed. As mentioned, bioreactor R2 was seeded with an inoculum obtained from bioreactor R1. Figures 6, 7 and 8 show the biogas production, chemical oxygen demand and pH responses, respectively, to disturbances P1, P2 and P3.

Figure 5. Acidogens: (1) Short and long rod-shaped bacteria, (2) diplococcus in chains (probably Streptococcus-like bacteria) and (3) filamentous bacilli.

Figure 6. Biogas response to disturbances P1, P2 and P3.

Figure 7. Effluent COD response to disturbances P1, P2 and P3.
High process efficiency values were obtained for all the disturbances applied. Indeed, efficiency of COD reduction varied between 93 and 98%. Disturbance P3 caused the lower system efficiency. The system pH remained self-regulated at the typical operation range of healthy methanogenic digesters (6.6-7.2) (Fig. 8). The biogas production rate showed the fastest (monitored) response to disturbances (Fig. 6). Chromatographic measurements of the composition of the gases accumulated in the gas collector varied between 83 and 88% for CH4, 15 and 10% for CO2 and about 2% for other gaseous components.
These results indicate that the microbial population present is able to resist increasing organic loading rate as a consequence of a mature and well-established methanogenic biofilm. The system seems to be capable of processing efficiently higher organic loadings than the here experienced. Unless the substrate fed to bioreactor R2 is mostly based on acetate, the milk powder supplement maintains active the acidogenic and acetogenic populations if a more complex substrate is to be treated.

Figure 8. pH response to disturbances P1, P2 and P3.
IV. CONCLUSIONS
The typical microbiological groups reported in literature under the experienced environmental conditions are present in the biofilm structure investigated on sand particles. This is concluded based on the morphology of the microorganisms analyzed by scanning electron microscopy. Methanosarcina sp. (acetate consumers) and filamentous microorganisms with morphology similar to Methanospirillum sp. (H2-utilizing archaea) can be distinguished among methanogens. Based on acetate levels and microorganism's threshold concentration, a predominance of Methanosarcina mazei rather than Methanosarcina barkeri is concluded. Among acetogens, bacillus morphologically similar to the syntrophic acetogenic Syntrophobacter wolinii can be observed together with Desulfovibrio sp. (hydrogen-utilizing, sulfate-reducing). With respect to acidogens, short and long rod-shaped bacteria, diplococcus in chains (probably Streptococcus-like bacteria) and filamentous bacilli can be morphologically distinguished but cannot be characterized from this study.
From a microbiological point of view, more specific analytical techniques are needed for a detailed characterization and evaluation of the ecological structure of the attached anaerobic consortia: direct chemotaxonomic methods like profiles of respiratory quinones (Hiraishi et al., 1989), inmunofluorescence (Lazarova and Manem, 1995; Amann et al., 1998), confocal scanning laser microscope (CSLM) (Davey and O'toole, 2000) and fluorescent in situ hybridization (FISH) (Großkopf et al., 1998; Davey and O'toole, 2000; Gonzalez-Gil et al., 2001; Díaz et al., 2003; Montenegro et al., 2003) can be mentioned.
A methanogenic biofilm fluidized bed reactor inoculated with the biofilm population investigated showed process efficiencies up to 98% for treating an acetate-based substrate. Based on this and on the other monitored variables, higher organic loading rates can a priori be processed without a serious detrimental effect on the process efficiency.
ACKNOWLEDGEMENTS
The financial support from the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), the Agencia Nacional para la Promoción de la Ciencia y la Tecnología (ANPCyT) and the Universidad Nacional del Litoral of Argentina is acknowledged.
REFERENCES
1. Amann, R., H. Lemmer and M. Wagner, "Monitoring the community structure of wastewater treatment plants: a comparison of old and new techniques", FEMS Microbiology Ecology, 25, 205-215 (1998). [ Links ]
2. Araujo, J.C., J.R. Campos and R.F. Vazoller. "Methanogenic biofilm: structure and microbial population activity in an anaerobic fluidized bed reactor treating synthetic wastewater", Biofilm Journal, 3, 1-14 (1998). [ Links ]
3. Batstone, D.J., J. Keller and L.L. Blackall, "The influence of substrate kinetics on the microbial community structure in granular anaerobic biomass", Water Research, 38, 1390-1404 (2004). [ Links ]
4. Bhatti, Z.I., K. Furukawa and M. Fujita, "Comparative composition and characteristics of methanogenic granular sludges treating industrial wastes under different conditions", Journal of Fermentation and Bioengineering, 79(3), 273-280 (1995). [ Links ]
5. de Bok, F.A.M., C.M. Plugge and A.J.M. Stams, "Interspecies electron transfer in methanogenic propionate degrading consortia", Water Research, 38, 1368-1375 (2004). [ Links ]
6. Boone, D.R. and M.P. Bryant, "Propionate-degrading bacterium, Syntrophobacter wolinii sp. nov. gen. nov., from Methanogenic Ecosystems", Applied and Environmental Microbiology, 40(3), 626-632 (1980). [ Links ]
7. Boone, D.R. and L. Xun, "Effects of pH, temperature, and nutrients on propionate degradation by a methanogenic enrichment culture", Applied and Environmental Microbiology, 53(7), 1589-1592 (1987). [ Links ]
8. Bramucci, M.G. and V. Nagarajan, "Industrial wastewater bioreactors: sources of novel microorganism for biotechnology", Trends in Biotechnology, 18, 501-505 (2000). [ Links ]
9. Brummeler, E., L.W. Hulshoff, J. Dolfing, G. Lettinga and A.J.B. Zehnder, "Methanogenesis in an upflow anaerobic sludge blanket reactor at pH 6 on an acetate-propionate mixture", Applied and Environmental Microbiology, 49(6), 1472-1477 (1985). [ Links ]
10. Davey, M.E. and G.O. O'toole, "Microbial biofilms: from ecology to molecular genetics", Microbiology and Molecular Biology Reviews, 64(4), 847-867 (2000). [ Links ]
11. Díaz, E., R. Amils and J.L. Sanz, "Molecular ecology of anaerobic granular sludge grown at different conditions", Water Science and Technology, 48, 57-64 (2003). [ Links ]
12. Ferry, J.G., P.H. Smith and R.S. Wolfe, "Methanospirillum, a new genus of methanogenic bacteria, and characterization of Methanospirillum hungatii sp. nov.", International Journal of Systematic Bacteriology, 24(4), 465-469 (1974). [ Links ]
13. Ghaly, A.E., "A comparative study of anaerobic digestion of acid cheese whey and dairy manure in a two-stage reactor", Bioresource Technology, 58, 61-72 (1996). [ Links ]
14. Gonzalez-Gil, G., P.N.L. Lens, A. Van Aelst, H. Van As, A.I. Versprille and G. Lettinga, "Cluster structure of anaerobic aggregates of an expanded granular sludge bed reactor", Applied and Environmental Microbiology, 67, 3683-3692 (2001). [ Links ]
15. Großkopf, R., P. Janssen and W. Liesack, "Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval", Applied Environmental Microbiology, 64(3), 960-969 (1998). [ Links ]
16. Hiraishi, A., K. Masamune and H. Kitamura, "Characterization of the bacterial population structure in an anaerobic-aerobic activated sludge system on the basis of respiratory quinone profiles" Applied and Environmental Microbiology, 55, 897-901 (1989). [ Links ]
17. Iannotti, E.L., J.R. Fisher and D.M. Sievers, "Characterization of bacteria from a swine manure digester", Applied and Environmental Microbiology, 43(1), 136-143 (1982). [ Links ]
18. Janssen, P.H., "Selective enrichment and purification of cultures of Methanosaeta spp.", Journal of Microbiological Methods, 52, 239-244 (2003). [ Links ]
19. Jetten, M.S.M., A.J.M. Stams and A.J.B. Zehnder, "Methanogenesis from acetate: a comparison of the acetate metabolism in Methanothrix soehngenii and Methanosarcina spp.", FEMS Microbiology Letters, 88(3-4), 181-197 (1992). [ Links ]
20. Lazarova, V. and J. Manem, "Biofilm characterization and activity analysis in water and wastewater treatment", Water Research, 29(10):2227-2245 (1995). [ Links ]
21. Le Mer, J. and P. Roger, "Production, oxidation, emission and consumption of methane by soils: A review", Eur. J. Soil Biol., 37, 25-50 (2001). [ Links ]
22. Lowe, S.E., M.K. Jain and J.G. Zeikus, "Biology, Ecology, and Biotechnological Applications of Anaerobic Bacteria Adapted to Environmental Stresses in Temperature, pH, Salinity, or Substrates", Microbiological Reviews, 57(2), 451-509 (1993). [ Links ]
23. McCabe, W.L., J.C. Smith and P. Harriott, Unit Operations of Chemical Engineering, McGraw-Hill Book Co, New York (1993). [ Links ]
24. Miyamoto, K., "Renewable biological systems for alternative sustainable energy production (FAO Agricultural Services Bulletin – 128)", FAO, Rome, Italy (1997). [ Links ]
25. Montenegro, M.A.P., J.C. Araujo and R.F. Vazoller, "Microbial community evaluation of anaerobic granular sludge from a hybrid reactor treating pentachlorophenol by using fluorescence in situ hybridization", Water Science and Technology, 48(6):65-73 (2003). [ Links ]
26. Perry, R.H. and C.H. Chilton, Chemical engineers' handbook, McGraw-Hill, New York (1973). [ Links ]
27. Speece, R.E., Anaerobic Biotechnology for Industrial Wastewaters, Archaea Press, Nashville, Tennesse (1996). [ Links ]
28. Stafford, D.A., D.L. Hawkes and R. Horton, Methane production from waste organic matter, CRC Press, Boca Raton, Florida (1980). [ Links ]
29. Westermann, P., B.K. Ahring and R.A. Mah, "Threshold acetate concentrations for acetate catabolism by aceticlastic methanogenic bacteria", Applied Environmental Microbiology, 55, 514-515 (1989). [ Links ]
30. Yang, S.T. and G.C. Guo, "A kinetic model for methanogenesis from whey permeate in a packed bed immobilized cell bioreactor", Biotechnology and Biochemistry, 37, 375-382 (1991). [ Links ]
31. Yang, Y., C. Tada, M.S. Miah, K. Tsukahara, T. Yagishita and S. Sawayama, "Influence of bed materials on methanogenic characteristics and immobilized microbes in anaerobic digester", Materials Science and Engineering C 24, 413-419 (2004). [ Links ]
32. Yu, H.Q., H. Fang and G.C. Guo, "Comparative performance of mesophilic and thermophilic acidogenic upflow reactors", Process Biochemistry, 38, 447-454 (2002). [ Links ]
33. Yu, J. and K.L. Pinder, "Intrinsic fermentation kinetics of lactose in acidogenic biofilm", Biotechnology and Bioengeenering, 41, 479-488 (1993). [ Links ]
34. Zellner, G., A.J.L. Macario and E.J.C. de Macario, "A study of three anaerobic methanogenic bioreactors reveals that syntrophs are diverse and different from reference organisms", FEMS Microbiology Ecology, 22, 295-301 (1997). [ Links ]
35. Zigová, J., E. Sturdík, D. Vandák and S. Schlosser, "Butyric acid production by Clostridium butyricum with integrated extraction and pertraction", Process Biochemistry, 34, 835-843 (1999). [ Links ]














