Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
Mastozoología neotropical
versión impresa ISSN 0327-9383versión On-line ISSN 1666-0536
Mastozool. neotrop. v.12 n.2 Mendoza jul./dic. 2005
Morphometric and allozymic characterization of Necromys benefactus populations in central Argentina
María C. Provensal1, Gladys E. Calderón2, Marina B. Chiappero3, Cristina N. Gardenal3, Jaime J. Polop1, and Marta S. Sabattini2
1 Departamento de Ciencias Naturales, Universidad Nacional de Río Cuarto, Río Cuarto, Córdoba, Argentina. cprovensal@exa.unrc.edu.ar. 2 Instituto Nacional de Enfermedades Virales Humanas, Pergamino, Argentina. 3 Cátedra de Genética de Poblaciones y Evolución, Fac. Cs. Exactas, Físicas y Naturales, Universidad Nacional de Córdoba, Córdoba, Argentina.
Key words. Genetic distance. Necromys. Morphometric variation. Oliveros virus. Pampa virus.
The genus Necromys Ameghino, 1889 (Rodentia, Sigmodontinae) is polytypic and widespread in central-eastern South America, comprising several different species distributed in the Central Andes, the Chacoan and Pampean regions and the northeastern Argentina, eastern and southern Brazil, and Uruguay (Reig, 1987; Galliari and Pardiñas, 2000).
Massoia and Fornes (1967) recognized two different species in the Pampean region and Uruguay; these authors argued that populations from southern Uruguay and southeastern Buenos Aires province correspond to Necromys obscurus and those in the north-west of Buenos Aires province belong to N. benefactus. Reig (1987) proposed that populations from Uruguay and southern Santa Fe, Córdoba, northern La Pampa, and northwestern Buenos Aires belong to the species N. obscurus, considering N. benefactus as a subspecies; those populations from the south-east and south-west of Buenos Aires province would represent a new species. Results of morphological and morphometric analysis by Galliari and Pardiñas (2000) reinforce the taxonomic and distributional hypothesis of Massoia and Fornes (1967) for the Pampean species of Necromys: N. obscurus would be restricted to southeastern Buenos Aires province and southern Uruguay and N. benefactus would have a broader distribution, from the south-east intermountain area of Buenos Aires province to the western and northeastern Buenos Aires, eastern of La Pampa, south of Santa Fe, and north of Córdoba province. No further information is available about distinctive morphometric and/or genetic characters useful to properly classify field specimens of these two species of Necromys.
It is well known that rodents are the most important natural reservoirs for zoonotic diseases (Hugh-Jones et al., 1995), although only a small proportion of the organisms infecting rodents would cause disease in humans. In the decade of 1990, a new arenavirus called Oliveros (OLV) was isolated from rodents captured at Oliveros and J.B. Molina (southern Santa Fe province), identified in the field as Bolomys obscurus (Mills et al., 1996) and then called N. benefactus (Galliari and Pardiñas, 2000). In 1992, a genomic variant of OLV denominated Pampa (PAM) was isolated from individuals captured at Maciel (southern Santa Fe province), apparently of the same rodent species. Serologic evidence of PAM infection in the rodent reservoir has been reported in several localities of the central region of Argentina (Riera, 2004). The pathogenesis of OLV virus is not known to date, but the emergence of this new infectious agent poses the need to improve our knowledge on biological properties of the host populations.
The precise determination of species boundaries and relationships among taxa in Sigmodontinae rodents is particularly difficult in some groups, mainly because of the poor morphological differentiation accompanying the speciation process in several genera (Steppan, 1993). In this paper, we characterize and compare individuals assigned to the genus Necromys trapped in different localities of the Pampean region of Argentina, using morphometric variables and allozymic frequencies.
The specimens of the genus Necromys were obtained at three localities: Oliveros (32°34'S, 60°51'W) and Uranga (33°16'S, 60°42'W) in Santa Fe province and Pergamino (33°32'S, 60°49'W) in Buenos Aires province (Fig. 1). Individuals were captured in the weedy borders of crop fields and roads with Sherman live traps set in lines of 125 m long, with one trap placed every 5 m. A preliminary taxonomic determination of Necromys individuals was made in the field on the basis of external features (size, fur coat and coloration) and total, tail, ear and right hind foot lengths. Twelve specimens from Uranga (5 males and 7 females) and 47 from Pergamino (22 males and 25 females) were carried alive to a field laboratory of the National Institute of Human Viral Diseases (Pergamino, Argentina) in order to establish two laboratory colonies. The animals were maintained under controlled conditions (18°- 22°C; 12L: 12D); food and water were provided ad libitum. Comparative morphometric studies were performed in these specimens and their descendants.

Fig. 1. Capture localities of Necromys specimens in the central-eastern Argentina.
Additional animals from Uranga (N = 11) and Pergamino (N = 20) and specimens trapped in Oliveros (N = 24) were sacrificed in the field, liver and kidneys were removed and preserved in liquid nitrogen for allozymic analyses.
Morphological studies: Specimens captured in Uranga and Pergamino were sacrificed after they left descendants. A second taxonomic determination was made then on these individuals, on the basis of qualitative cranial characters described by Galliari and Pardiñas (2000): nasals flat, nasofrontal suture with respect to the orbital process, frontal edges, interorbital constriction, free upper border of zygomatic plate, shape of the mesopterygoid fossa, and direction of the coronoid process in relation to articular process and develop of the coronoid process.
One hundred and forty four specimens of > 60 days old from the first generation of both colonies were compared separately by sex, considering the results of a previous study of morphometric characters in animals of different age classes (Polop et al., 2000). The animals were sacrificed by inhalation of anesthetic gas (Methoxyflurane), and the following characters measured: body weight (BW), nasal width (NW), length of hard palate (PL), length of upper diastema (D), mandible length (ML), zygomatic width (ZW), and nasal length (NL). Body weight was recorded to the nearest 0.5 g in animals 60 to 120 days old, and the arithmetic mean for each group (according to sex and location) was compared. Measurements were taken with caliper and recorded to the nearest 0.02 mm.
The similarity between colonies and between adults from Uranga and Pergamino were established using the D2 of Mahalanobis (Mahalanobis, 1936; Rao, 1952). A cluster analysis was performed by the UPGMA procedure, based on pairwise Euclidean distances among individuals from all colonies and sites (male and female individuals analyzed separately).
Allozymic studies: In order to examine the degree of similarity in the gene pool of the populations assigned to the species of genus Necromys, we analysed the allozymic polymorphism in individuals captured in Oliveros (N=24), Uranga (N=11) and Pergamino (N=20). Twelve informative loci from 9 enzymatic proteins were studied: lactate dehydrogenase (Ldh, E.C. 1.1.1.27), leucine aminopeptidase (Lap, E.C. 3.4.11), malic enzyme (ME, E.C. 1.1.1.40), α-glycerophosphate dehydrogenase (Gpdh, E.C. 1.1.1.8), esterases (Est-5 and Est-6, E.C. 3.1.1.1), kidney and liver acid phosphatases (AcpK -1, AcpK -2 and AcpL, E.C. 3.1.3.2), glutamate oxaloacetate transaminase (Got-1, E.C. 2.6.1.1), glucose-6-phosphate dehydrogenase (G6pdh, E.C. 1.1.1.49) and 6-phosphogluconate dehydrogenase (6-PGD, E.C. 1.1.1.44). Preparation of homogenates, electrophoresis and staining to reveal enzyme activity was performed as described by Selander et al. (1971), and Gardenal and Blanco (1985).
Deviations from Hardy-Weinberg equilibrium in each locus in each population were tested by the exact test based on a Markov chain algorithm (Guo and Thompson, 1992) using GENEPOP version 3.2a (Raymond and Rousset, 2000). Genetic structure of populations was estimated by F-statistics, using the unbiased estimators of Weir and Cockerham (1984): Θ for FST and f for FIS. Significance of Θ and f values were obtained by randomization procedures, as implemented in FSTAT program (Goudet, 2000). If there are two cryptic species, each population sample will contain individuals pertaining to two separate gene pools and f value will be significant and positive. If each sample comes from a single panmictic population, f will not be statistically different from 0.
Mean heterozygosity per locus, percentage of polymorphic loci (P95% criterion) and mean number of alleles per locus (A) were calculated using BIOSYS-2 (Black, 1997).
Specimens captured in the different localities presented the following characteristics with respect to the fur coat and coloration: the dorsum had short guard hairs (9- 10 mm), with a yellowish pheomelanin portion, giving the back an agouti colour. The sides of the body were yellowish-grey and the abdomen was whitish grey, with white gular and inguinal regions. The range of the length of head and body was 135- 209 mm and the range of the length of tail was 48- 84 mm. These features correspond with the description of Necromys benefactus made by Galliari and Pardiñas (2000).
Considering the morphological studies all the specimens captured in Uranga, Pergamino and those obtained from the laboratory colonies showed the following states of qualitative characters: a) nasal projected rostrally, overhanging the premaxillae and hiding the incisors in dorsal view; b) conspicuous interorbital constriction, frontals divergent caudally, with crested and sharp lateral edges; c) zygomatic plate high, not robust, with its dorsal margin shorter and slightly rounded in its antero-dorsal end; d) mesopterygoid fossa squared off, with a well-defined medial palatine process of consistent occurrence across individuals; e) a long and slender coronoid process, with its end completely curved caudally. These character states corresponded to those that diagnose Necromys benefactus (Galliari y Pardiñas, 2000).
The descriptive statistics for mophometric data of the two colonies are shown in Table 1.

The comparison between colonies showed a value of D 2 = 0.632 (F=0.937; d.f.7 and 67; p=0.48) for females and 1.33 (F=0.8; d.f.7 and 61; p=0.60) for males. The groups formed when the UPGMA method was applied to similarity data, included individuals from both colonies (Figs. 2a and 2b).
Fig. 2. Unweighted pair-group average clustering of Euclidean distances among individuals of Necromys benefactus from Pergamino (p) and Uranga (u) colonies. a) Males. b) Females.
The low values of distance, the non significant results of F statistic and the cluster analysis reflect minimal variation between individuals from the colonies. Similar results were obtained with individuals trapped in the field (Fig. 3a and 3b).
Fig. 3. Unweighted pair-group average clustering of Euclidean distances among adults of Necromys benefactus trapped in field (p = Pergamino and u = Uranga). a) Males. b) Females.
The results for the allozymic studies show that in the three populations of Necromys studied (Oliveros, Uranga and Pergamino), observed genotypic frequencies did not deviate from those expected under Hardy Weinberg equilibrium.
Four out of 16 loci (Gpdh, Got-1, G6pdh and 6Pgdh) were monomorphic in the three populations analyzed. Allele frequencies for polymorphic loci are given in Table 2. Percentage of polymorphic loci and average heterozygosity are shown in Table 3.


Nei's genetic distance between pairs of populations were the following: Oliveros-Pergamino, D = 0.007; Oliveros-Uranga, D = 0.008 and Pergamino-Uranga, D = 0.02.
In agreement with the non significant deviations from random mating, f values for individual loci were not significantly different from 0 (Table 4). Mean theta value was 0.027, which was significant at the 5% level.
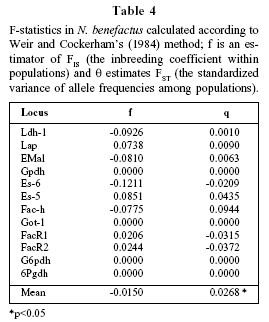
The comparative analysis of external characters and qualitative traits of the skull in individuals from two localities of the Pampean region, allowed us to assign all the specimens analyzed to N. benefactus, according to the nomenclature proposed by Galliari and Pardiñas (2000).
A statistical analysis of morphometric parameters between individuals captured in the field as well as their descendants obtained in laboratory colonies, did not show significant differentiation according to their sampling site. Both males and females presented high similarity levels in mean values of six craniomorphic measures and body weight; none of the variables tested was useful to discriminate individuals by their geographic origin. Clustering of individuals also showed that specimens from Uranga and Pergamino do not form separate groups. Galliari and Pardiñas (2000) observed that populations of N. benefactus from northeastern Buenos Aires Province (coastal grasslands) and those from Sierra de la Ventana (a highland area in southeastern Buenos Aires) showed morphometric differentiation, which could justify their sub-specific separation. Our results suggest, on the basis of D2 values, that populations from the two locations studied were not differentiated at the subspecific level.
The absence of significant morphometric variation is in agreement with the results from allozymic studies. Observed genotype frequencies in specimens captured at each location conform to the Hardy-Weinberg equilibrium. No fixed exclusive alleles for any of the populations were detected and Nei's genetic distance values among Uranga, Pergamino and Oliveros populations were very low (between 0.007 and 0.02), confirming that the rodents sampled belong to a single species. Although the small but statistically significant value of q indicates a moderate degree of genetic subdivision between populations, it is similar to the values reported for other sigmodontine species living in the same area: Calomys musculinus (Chiappero et al., 2002a), C. laucha (Gardenal et al., 2002; Chiappero et al., 2002 b) and Oligoryzomys flavescens (Chiappero et al., 1997).
Levels of genetic variability found in the populations studied (P95% between 41.7 and 58.3%; H between 0.111 and 0.148) were considerably higher than those obtained by Apfelbaum and Reig (1989) in a population from southern Buenos Aires province of specimens classified as N. obscurus benefactus (P95% =10.5; H=0.024). However, the levels of polymorphism of our study were close to those observed in other sigmodontine species from the Pampean region (vide supra).
Lozano et al. (1997) reported that the nucleotide sequence identity between the viral forms OLV and PAM was 84.6%, which would fall very close to the proposed cut off between species and strains for other arenavirus genotypes (García et al., 2000). According to our results, the same species of Necromys would be present in Oliveros, Pergamino and Uranga, with low to moderate genetic differentiation among populations. Because the 3 rodent populations belong to the same species, specificity of host cannot be used as evidence that the two viral forms are different species. However, the possibility that they were actually different viruses cannot be ruled out until more detailed genetic studies are performed.
We thank to field assistants of the INEVH for the specimens provided. This work was partially founded by Roemmers Fundation, the Agencia Córdoba Ciencia and the SECYT, Universidad Nacional de Córdoba, Argentina.
LITERATURE CITED
APFELBAUM LI and OA REIG. 1989. Allozyme genetic distances and evolutionary relationships in species of akodontine rodents (Cricetidae: Sigmodontinae). Biological Journal of the Linnean Society 38:257-280. [ Links ]
BLACK IV WC. 1997. BIOSYS-2: a computer program for the analysis of allelic variation in Genetics. Available in: http//www.lamar.colastate.edu. [ Links ]
CHIAPPERO MB, A BLANCO, GE CALDERÓN, MS SABATTINI, and CN GARDENAL. 2002b. Genetic structure of populations of Calomys laucha (Muridae, Sigmodontinae) from central Argentina. Biochemical Systematics and Ecology 30:1023-1036. [ Links ]
CHIAPPERO MB, GE CALDERÓN and CN GARDENAL. 1997. Oligoryzomys flavescens (Rodentia, Muridae): gene flow among populations from Central- eastern Argentina. Genetica 101:105-113. [ Links ]
CHIAPPERO MB, MS SABATTINI, A BLANCO, GE CALDERÓN and CN GARDENAL. 2002a. Gene flow among Calomys musculinus (Rodentia, Muridae) populations in Argentina. Genética 114:63-72. [ Links ]
GALLIARI CA and UFJ PARDIÑAS. 2000. Taxonomy and distribution of the sigmodontine rodents of genus Necromys in central Argentina and Uruguay. Acta Theriologica 45:211-232. [ Links ]
GARCÍA JHB, SP MORZUNOV, S LEVIS, G ROWE, G CALDERÓN, D ENRÍA, MS SABATTINI, MJ BUCHMEIER, MD BOWEN and ST JEOR. 2000. Genetic diversity of the Junin virus in Argentina: geographic and temporal patterns. Virology 272:127-136. [ Links ]
GARDENAL CN, MB CHIAPPERO, GM DE LUCA D'ORO and JN MILLS. 2002. Allozymic polymorphism and genetic differentiation among populations of Calomys musculinus and Calomys laucha (Rodentia: Muridae) from eastern Argentina. Mammalian Biology 67:294-303. [ Links ]
GARDENAL CN and A BLANCO. 1985. Polimorfismo enzimático en Calomys musculinus: nueva estimación. Mendeliana 7:3-12. [ Links ]
GOUDET J. 2000. FSTAT, a program to estimate and test gene diversities and fixation Indices (version 2.9.1). Available in: http://www.unil.ch/izea/softwares/fstat.html [ Links ]
GUO S and E THOMPSON. 1992. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48:361-372. [ Links ]
HUGH-JONES ME, HUBBERT WT and HV HAGSTAD. 1995. Zoonoses: recognition, control and prevention. Iowa State University press, Ames. [ Links ]
LOZANO ME, DM POSIK, CG ALBARIÑO, G SCHUJMAN, PD GHIRINGHELLI, GE CALDERÓN, MS SABATTINI and V ROMANOWSKI. 1997. Characterization of arenaviruses using a family-specific primer set for RT-PCR amplification and RFLP analysis. Its potential use for detection of uncharacterized arenaviruses. Virus Resarch 49:79-89. [ Links ]
MAHALANOBIS PC. 1936. On the Generalized Distance in Statistics. National Institute of Science India 2:49-55. [ Links ]
MASSOIA E and A FORNES. 1967. El estado sistemático, distribución geográfica y datos etoecológicos de algunos mamíferos neotropicales (Marsupialia y Rodentia) con la descripción de Cabreramys, género nuevo (Cricetidae). Acta Zoológica Lilloana 23:407-430. [ Links ]
MILLS JN, JG BARRERA ORO, DS BRESSLER, JE CHILDS, RB TESH, JF SMITH, DA ENRIA, TW GEISBERT, KT McKEE JR., MD BOWEN, CJ PETERS and PB JAHRLING. 1996. Characterization of Oliveros Virus, an new member of the Tacaribe Complex (Arenaviridae: Arenavirus). American Journal Tropical Medicine Hygiene 54:399-404. [ Links ]
POLOP JJ, J LISANTI, EA CASTILLO, MC PROVENSAL, JB GARCÍA, A MORALES, MI ORTIZ and MS SABATTINI. 2000. Estudio de la patogenicidad de los nuevos arenavirus Oliveros y Pampa y de sus hospederos del género Bolomys (Rodentia, Muridae). Anales de la Fundación Alberto J. Roeemers 13: 473-504. [ Links ]
RAYMOND M and F ROUSSET. 2000. GENEPOP 3.2a version. Available in: http://www.cefe.cnrs-mop.fr. [ Links ]
RAO CR. 1952. Advanced statistical methods in biometrics research. J. Willey, New York. [ Links ]
REIG OA. 1987. An assessment of the systematic and evolution of the Akodontini, with the description of a new fossil species of Akodon (Cricetidae: Sigmodontinae). Fieldiana: Zoology (new series) 39:347-399. [ Links ]
RIERA LM. 2004. Arenavirus que coexisten con el virus Junin (VJ) en la Pampa Argentina: su asociación con patologías humanas. PhD thesis. Universidad Nacional del Litoral. [ Links ]
SELANDER RK, MH SMITH, SY YANG, WE JOHNSON and JB GENTRY. 1971. Studies in genetics. VI. Biochemical polymorphism and systematics in the genus Peromyscus. I. Variation in the old-field mouse (Peromyscus polionotus): Universidad de Texas Publicaciones 7103:49-90. [ Links ]
STEPPAN SJ. 1993. Phylogenetic relationships among the Phyllotini (Rodentia: Sigmodontinae) using morphological characters. Journal of Mammalian Evolution 1:187-213. [ Links ]
WEIR BS and CC COCKERHAM. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38:1248-1370. [ Links ]
Recibido 4 febrero 2005.
Aceptación final 24 junio 2005.














