Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
Mastozoología neotropical
versión impresa ISSN 0327-9383
Mastozool. neotrop. vol.17 no.2 Mendoza dic. 2010
ARTÍCULOS
A Molecular Perspective On The Diversification Of Short-Tailed Opossums (Monodelphis: Didelphidae)
Sergio Solari
Department of Biological Sciences, Texas Tech University, Lubbock, TX 79409, USA. Current address: Instituto de Biología, Universidad de Antioquia, A.A. 1226, Medellín, Colombia <ssolari@matematicas.udea.edu.co>
Recibido 6 enero 2009.
Aceptado 15 setiembre 2010.
Editor asociado: G D'Elía
ABSTRACT: As currently understood Monodelphis includes more than 22 species and is the most diverse genus of opossums (Didelphimorphia). No complete evaluation of the systematic relationships of its species has been attempted, despite the fact that several species groups and even genus-level groups have been proposed based on morphology, and that some of them are limited to recognized biogeographic regions. Here, genealogic relationships among 17 species were assessed based on phylogenetic analyses of 60 individual sequences (801 base pairs of the mitochondrial cytochrome b gene). The analyses cast doubts on the monophyly of Monodelphis, but species were consistently discriminated into eight species groups: (a) brevicaudata group, with five species, (b) adusta group, four species, (c) dimidiata group, two species, (d) theresa group, two species, (e) emiliae group, (f) kunsi group, (g) americana group, and (h) 'species C' group; the last four being monotypic. Genetic divergence among species within species groups ranges from 0.48 to 12.27%, and among species groups it goes from 15.61 to 20.15%. Further analyses in some species groups reveal congruence between genetic divergence, morphological traits, and geographic distribution, providing additional support for recognition of species limits. Although the cytochrome b gene may diverge too fast to evaluate relationships among the older lineages of the genus, the use of a broad taxon sampling allows for independent tests of hypotheses on species limits and relationships based on non-molecular characters. Congruent patterns offer a starting point for developing a sound taxonomy for Monodelphis and more robust hypotheses in regard to its diversification over many diverse Neotropical habitats.
RESUMEN: Una perspectiva molecular sobre la diversificación de las comadrejas colicortas (Monodelphis: Didelphidae). La definición actual del genero Monodelphis incluye más de 22 especies, siendo el género más diverso de marsupiales (Didelphimorphia). Una evaluación de las relaciones sistemáticas entre especies no ha sido intentada aún, a pesar de haberse propuesto, en base a morfología, varios grupos de especies e incluso nombres del nivel de género, y que algunos de ellos parecen limitados a regiones biogeográficas reconocidas. Las relaciones genealógicas entre 17 especies fueron evaluadas en base a análisis filogenéticos de 60 secuencias individuales (801 pares de bases del gen mitocondrial Citocromo b). Aunque los análisis no corroboran la monofilia de Monodelphis, las especies fueron discriminadas consistentemente en ocho grupos de especies: (a) grupo brevicaudata, con cinco especies, (b) grupo adusta, cuatro especies, (c) grupo dimidiata, dos especies, (d) grupo theresa, dos especies, (e) grupo emiliae, (f) grupo kunsi, (g) grupo americana, y (h) grupo 'especie C'; siendo los cuatro últimos monotípicos. La divergencia genética entre especies del mismo grupo varía entre 0.48 y 12.27%, y entre grupos varía de 15.61 a 20.15%. Análisis más detallados en algunos grupos muestran congruencia entre patrones de variación molecular, características morfológicas y distribución geográfica, brindando soporte adicional a las relaciones encontradas. Aunque el gen citocromo b puede divergir demasiado rápido para evaluar relaciones entre grupos antiguos en este género, un muestreo taxonómico amplio y denso permite una prueba independiente de hipótesis basadas en caracteres no moleculares sobre límites entre especies y sus relaciones. Estos patrones congruentes ofrecen un punto de partida para el desarrollo de una taxonomía válida en Monodelphis e hipótesis más robustas respecto a su diversificación a través de los diversos hábitats del Neotrópico.
Key words. Didelphimorphia; Phylogenetics; Species limits; Taxonomy.
Palabras claves. Didelphimorphia; Filogenética; Límites entre especies; Taxonomía.
INTRODUCTION
The genus Monodelphis includes small to mediumsized shorttailed opossums found throughout various habitats from southeastern Panama to northeastern Argentina, ranging from near sea level up into montane forests (Solari, 2007). Although it is the most diverse genus of living didelphids, only five other genus-level names have been proposed to group species, but none is currently treated as a valid subgenus (Gardner, 2005; Pine and Handley, 2008). A number of species groups have been recognized in the literature (e.g., Matschie, 1916; Gilmore, 1941; Pine, 1976; Gomes, 1991), and sometimes allocated to the genus-level names, but their definition and limits have been inconsistent. Most of the groups are based on a few conspicuous external characters, the most obvious being fur coloration of the back and sides. Additional details concerning skulls and teeth appear in a few diagnoses, while occasionally species are aggregated based on geographic grounds (e.g., Solari, 2004).
Aside from the conspicuously short tail common to all species, other evidence suggesting a monophyletic Monodelphis is a diploid number of 2N=18, unique among marsupials (Reig et al., 1977). Preliminary and sparse data on chromosomes (Reig et al., 1977; Langguth and Lima, 1988; Palma and Yates, 1996; Carvalho et al., 2002) show some diversity worthy of further study, but no one has proposed relationships based on that dataset. Currently, based on morphological and genetic data, the genus is considered monophyletic (Voss and
Jansa, 2003, 2009). Hypotheses concerning relationships have been advanced for some species, based on morphometric (Ventura et al., 1998) or genetic data such as mitochondrial (Patton and Costa, 2003; Solari, 2007) and nuclear genes (Voss and Jansa, 2003, 2009). The most extensive (but unpublished) review of the genus is that of Gomes (1991), with emphasis on species from Brazil; however, that taxonomy disagrees with the one used by most current authors (e.g., Gardner, 2005; Pine and Handley, 2008). The taxonomy of the genus and species has been updated by Pine and Handley (2008), summarizing their previous findings, as well as those by Ventura et al. (1998), Voss et al. (2001), and Solari (2007). These studies have provided valuable insight into the systematics of the genus, but the clear existence of species complexes raises questions about the number of species and their actual relationships.
This study examines the diversity and phylogenetic relationships among several species of Monodelphis. Given our limited knowledge of the relationships within Monodelphis, and that so far no study has included a representative subset of species, wide taxonomic and geographic sampling is imperative; as such, emphasis is given to obtaining a broad representation of taxa and collection localities. This dataset includes 14 currently recognized species plus three recognized as distinct but unnamed forms (species B, species C, and species D; see Pine and Handley, 2008). Mitochondrial sequences (cytochrome b gene) are used to provide a genealogical framework for assessing species limits and to address the existence of species groups within the genus, and if any of these correspond to groups previously identified.
MATERIALS AND METHODS
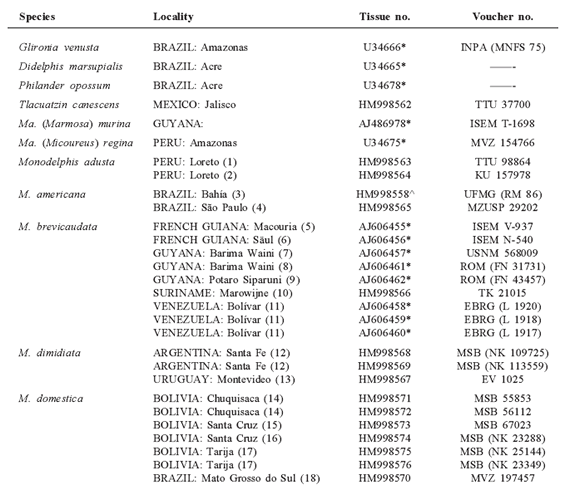
Samples. Forty one tissues samples representing 15 Monodelphis species, one tissue sample of Tlacuatzin (part of the outgroup), plus 25 cytochrome b sequences (including five Monodelphis species and five of the outgroup species) directly obtained from GenBank (Appendix) were used in this research. Collecting localities and museum catalog [or collector's field] numbers are given for vouchers and tissues, or at least for whichever one was available at the time of analysis (Appendix); the localities are mapped in Fig. 1. Voucher specimens are housed in the following institutions: American Museum of Natural History (AMNH, New York), Colección Boliviana de Fauna (CBF, La Paz, Bolivia), Laboratorio de Evolución, Universidad de la República (EV, Montevideo, Uruguay), Field Museum (FMNH, Chicago), University of Kansas Museum of Natural History (KU, Lawrence), Museum of Southwestern Biology (MSB, Albuquerque), Museo de Historia Natural, Universidad Nacional Mayor de San Marcos (MUSM, Lima, Peru), Museum of Vertebrate Zoology (MVZ, Berkeley), Natural Sciences Research Lab, The Museum, Texas Tech University (TTU, Lubbock), and National Museum of Natural History (USNM, Washington, D.C.). Some Brazilian specimens sequenced by J. L. Patton and collaborators are deposited in the following institutions: Instituto Nacional de Pesquisas da Amazonia (INPA, Manaus), Museu Paraense Emilio Goeldi (MPEG, Belém), Museu de Zoologia, Universidade de São Paulo (MZUSP, São Paulo), and Universidade Federal de Minas Gerais (UFMG, Belo Horizonte).

Fig. 1. Collection localities of the specimens included in the present analysis. To improve resolution, samples are arranged according to the resulting groups (see included species in text). Locality records correspond to brevicaudata group [triangles], adusta group [circles], dimidiata group [diamonds], theresa group [crosses], emiliae complex [asterisks], kunsi complex [stars], americana complex [squares], and 'species C complex [encircled dots]. Numbers beside the symbols make reference to the localities as detailed in Appendix 1. Country ñames as follows: AR=Argentina, BO=Bolivia, BR=Brazil, CL=Chile, CO=Colombia, EC=Ecuador, GF=French Guiana, GY=Guyana, PE=Peru, PY=Paraguay, SR=Suriname, UY=Uruguay, VE=Venezuela.
DNA sequence acquisition. Total genomic DNA was extracted following available protocols based on phenolchloroform, proteinase K-ribonuclease (Longmire et al., 1997; Sambrook et al., 1989), or manufacturer's protocol of the DNeasy tissue kit (Qiagen Inc., Valencia, CA). Double-stranded amplification of the entire cytochrome b gene was performed via polymerase chain reaction (PCR), using primers MVZ05 and MVZ14 (Patton et al., 1996). Conditions for these reactions were: 35 cycles of 94° C for 50 seconds, 47°C for 1 minute, and 72°C for 1.5 minutes; the cycle was then held at 72°C for 30 minutes to allow for extension of DNA. PCR products were then purified using a Qiagen PCR purification kit (Qiagen Inc.) following the instructions of the manufacturer. Amplicons were sequenced using ABI Big Dye Terminator Ready Reaction Mix, following manufacturer's instructions. Eight primers aimed to obtain the whole gene were used in the sequencing protocol: two (MVZ05 and MVZ14) used in the amplification, three (MVZ04, MVZ11, and MVZ16) used by Patton et al. (1996) and Patton and Costa (2003), and three (mono700H1, mono480L, and mono860L) specifically designed for Monodelphis (Solari, 2007). DNA was sequenced in both directions to ensure accuracy of generated sequences in an ABI Prism 377 Genetic Analyzer (Perkin Elmer, Applied Biosystems, Foster City, CA).
Phylogenetic analyses. Sequence alignment was performed using Vector NTI Suite 6.0 (InforMax Inc, Bethesda, MD), specifically ContigExpress (for DNA sequence assembly), and Align X (for múltiple alignment of sequences) components. Three different analyses: máximum parsimony, máximum likelihood, and Bayesian were performed using PAUP* 4.0b 10 (Swofford, 2002) and MrBayes 2.01 (Huelsenbeck and Ronquist, 2001). Topologies were rooted using an outgroup formed by six species: Glironia venusta, Didelphis marsupialis, Philander opossum, Tlacuatzin canescens, Marmosa (Marmosa) murina, and M. (Micoureus) regina. Genera Tlacuatzin and Marmosa, along with Monodelphis, have been included in the tribe Marmosini Hershkovitz 1992 by Voss and Jansa (2009). For Máximum Parsimony (MP) analysis, nucleotide positions were treated as discrete, unordered, and equally weighted characters with four character states (A, C, G, and T). Trees were found by heuristic search with 100 random sequence addition, and treebisection-reconnection (TBR) branch swapping. When múltiple minimum-length trees were found, strict consensus trees were obtained. Máximum Likelihood (ML) trees were calculated based on the selection of the most appropriate model of DNA sequence evolution as determined by Modeltest (Posada and Crandall, 1998). The GTR + r+ /model generated significantly better likelihood scores and therefore was used in all likelihood analyses. This model included a proportion of invariable sites and a gamma shape parameter. Analyses employed empirical base compositional biases, 50 random input orders, and TBR branch swapping. Reliability of nodes was evaluated through bootstrap analyses (Felsenstein, 1985) with 200 iterations in the full-heuristic mode (5 random-additions) for the MP topology, and 1000 iterations in fast mode for the ML topology.
For Bayesian analyses, the GTR model of evolution, with a gamma parameter and a proportion of invariable sites, was used. Two runs were conducted simultaneously, each with four Markov chains with random starting trees ran for 1 000 000 generations; trees were sampled every lOOth generation, and 'burnin' valúes determined by empirical evaluation of likelihood scores. A majorityrule consensus tree was calculated from the sample of stabilized trees in PAUP*, with branch lengths obtained via the 'sumt' option in MrBayes. Clade reliability was assessed via posterior probabilities, and valúes > 0.95 regarded as significant.
Genetic divergence among species and clades was estimated from Kimura-2 parameter distances as implemented in PAUP*. This specific model of evolution allows direct comparison with previous studies involving short-tailed opossums (Patton and Costa, 2003; Steiner and Catzeflis, 2004; Solari, 2007), as well as to estimate minimum divergence between recognizable sister taxa (Avise, 2004; Baker and Bradley, 2006), thus helping to under-stand morphological and geographic diversification. It will allow testing the phylogenetic value of previous taxonomic arrangements, such as species groups or genus-level groups, as well as geographic hypotheses of diversification (e.g., Patton et al., 2000; Costa, 2003) in particularly wellsampled groups of species.
RESULTS
Partial sequences (ranging from 790 to 830 bp) were obtained from 17 species (60 individuals examined) of Monodelphis; 41 of these correspond to new sequences (GenBank accession numbers HM998558-998598). Analyses of the first 801 bp dataset, including the outgroup genera, resulted in 448 constant and 309 parsimony informative characters. Average nucleotide frequencies of informative characters were: adenine 0.33, cytosine 0.32, guanine 0.04, and thymine 0.31. Among these, 58 (19%) were 1st position changes, 14 (5%) were 2nd position, and 237 (76%) were 3rd position; this proportion is concordant with cytochrome b variation in opossums (Patton et al., 1996; Mustrangi and Patton, 1997). Likewise, saturation values indicate that third base positions reached saturation point for transitions at around 15% sequence divergence, with a possible loss of phylogenetic signal among more divergent taxa in the analysis.
The MP analysis generated 240 most parsimonious trees, each 1835 steps long, with a CI of 0.30 and a RI of 0.75; the consensus tree (not shown) recovered eight major lineages (thereafter, "species groups") with high nodal bootstrap support (Table 1); these groups are referred as to adusta, americana, brevicaudata, dimidiata, emiliae, kunsi, theresa, and one unnamed species group. Relationships among the species groups are unresolved. However, Monodelphis did not appear as monophyletic because a clade with both species of Marmosa (subgenera Marmosa and Micoureus) is nested within these eight species groups.
Table 1 Support values for specific nodes in each of the three phylogenetic analyses (bootstrap values [BS] are provided for MP and ML trees and posterior probabilities [PP] for Bayesian analyses) of different species of short-tailed opossums of the genus Monodelphis. Nodes are identified numerically as in the topology of Fig. 2.
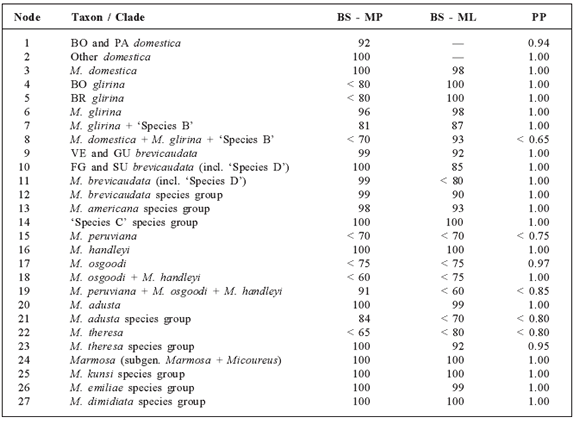
The máximum likelihood analysis used the GTR + r+ /model of evolution with gamma shape parameter = 1.2086, and proportion of invariable sites = 0.5383. The topology (Fig. 2; -ln L = 8592.478) recovered the eight species groups of the MP analyses, all with BS > 90 (except for the adusta group; Table 1); however, the genus results polyphyletic by inclusión of both species of Marmosa as sister to the theresa species group, but this relationship had low nodal support (BS < 50). Likewise, deeper nodes and relationships among lineages had low support (BS < 65). Bayesian analysis recovered a topology (not shown) similar to that of the ML, with minimal differences related to the position of some terminals within species.

Fig. 2. Optimal tree obtained by Máximum Likelihood analysis (-ln L=8592.478) based on cytochrome b gene sequences of several species of Monodelphis using the GTR +T+ /model of molecular evolution. Species groups are labeled as in text. See Table 1 for bootstrap support valúes (form MP and ML trees), and Bayesian posterior probabilities corresponding to each labeled branches. Nodes marked with an asterisk (*) liad low support valúes (BS < 50; PP <0.7) in all the analyses.
Genetic divergence based on the Kimura 2-parameters model ranged from 0.4 to 20.6% among species of Monodelphis. Average internal divergence within species groups ranged from 0.48% (dimidiata species group) to 12.27% (adusta species group); pairwise divergence had the highest value within the brevicaudata species group, with 13.9% between brevicaudata and glirina. Divergence between taxa in different species groups ranged from 15.8% (americana vs. species C) to 20.6% (domestica vs. emiliae). Table 2 lists genetic divergence values within and among these eight species groups and with Tlacuatzin, the closest outgroup genus that stayed outside Monodelphis.
Table 2 Genetic divergence (Kimura 2-parameter distance) within [along the diagonal] and among [below the diagonal] the eight species groups of Monodelphis species identified in the molecular analyses of cytochrome b gene sequences. Tlacuatzin, one of the genera used as outgroup, is included for comparisons.

DISCUSSION
Results of maximum parsimony, maximum likelihood, and Bayesian analyses produced trees with slightly different topologies, although all depicting eight distinctive, consistent and well supported species groups (Table 1); four correspond to lineages composed by two or more species and the other four are monospecific. Due to this overall similarity, the maximum likelihood tree (Fig. 2) was chosen for the following discussion. Relationships among species groups did not receive enough support to allow further discussion of deeper nodes.
Monophyly of the genus Monodelphis
None of these analyses recovered a monophyletic Monodelphis. Even with a large taxonomic sampling of the genus (17 of 22-23 species recognized by recent revisions; Solari, 2007; Pine and Handley, 2008), parsimony and model based analyses failed to recover a monophyletic Monodelphis. Although some morphological traits and its unique chromosome number have been used to define the genus (Creighton, 1984; Reig et al., 1987), most of this evidence needs to be reassessed within a phylogenetic context. In a similar way, the morphological characters proposed by Goin and Rey (1997) do not apply to all Monodelphis species (pers. obs.). The most complete morphological dataset for didelphids (Voss and Jansa, 2003, 2009) identified a single discrete character state (maxilloturbinals simple, slender, and unbranched) as a synapomorphy for the four Monodelphis species (brevicaudata, emiliae, peruviana, theresa) assessed in the analysis. A reassessment of these putative synapomorphies needs to be completed for a larger set of species. Voss and Jansa (2009) also found a well-supported monophyletic genus based on independent and combined analyses of five gene sequences. Therefore, although the monophyly of Monodelphis was not clearly supported in the present molecular analysis, keeping this as a working hypothesis seems appropriate (see below).
Indiscriminate use of the cytochrome b gene as a genetic marker is criticized because of faster rate of mutation and high saturation (Jansa and Voss, 2000; Springer et al., 2001), which result in an inability to resolve deep divergences. Age estimation based on molecular dating of nuclear genes (Steiner et al., 2005) points to a Monodelphis origin as early as the late Oligocene (26.1 ± 3.3 Mya). Therefore, it is possible that the cytochrome b could be useful in solving relationships of recently diverged species (see Mustrangi and Patton, 1997; Patton and da Silva, 1997; Costa et al., 2003; Solari, 2007), but not deeper at or near the base of the genus. Therefore, the inclusion of more slowly nuclear sequences is needed to further test the monophyly of Monodelphis and relationships among the eight species groups.
Definition of species and species group
Within the ML tree, eight (americana, adusta, domestica, emiliae, glirina, handleyi, kunsi, and species C) out of 17 putative species received high support (BS > 90, PP > 0.95), whereas osgoodi had PP > 0.95 but BS < 85 and peruviana had low support. Three putative species (scalops, sorex, and species D) appeared nested within others (theresa, dimidiata, and brevicaudata, respectively) failing the criterion of reciprocal monophyly for specific distinction. Additional studies should be conducted to resolve their status as species. Finally, one undescribed species ('species B', in Pine and Handley, 2008) was represented by a single specimen and its support cannot be established. Genetic divergence among species ranged widely, 0.48-20.6%, depending on the scale of comparison; it was lower within species groups and larger between species from different species groups. This variation makes possible to use the lowest divergence between fully recognized sister species as a threshold useful to tentatively indicate specific level of lineages within some of these species that should be further tested with other analyses; for instance, within the adusta and the brevicaudata species groups the average divergence between sister species was 10%. Therefore, given that sequence divergence between sister taxa M. glirina and 'species B' was 8.65%, and the latter has been morphologically considered similar to M. domestica (see Pine and Handley, 2008) these data are indicative of specific status for 'species B'. The implications of these criteria for the recognition of Monodelphis species is further discussed below.
It is worthy of mention that four well-supported species groups were corroborated by different tree-building methods. Species groups have been recognized in Monodelphis and even given genus-group names (e.g., Matschie, 1916; Cabrera, 1919), but this study constitutes the first test of their monophyly. However, the fact that the phylogenetic analyses lack a number of species must be considered, because their inclusion may affect the composition and limits of some of these groups. Support of the eight species groups in all the phylogenetic analyses is remarkable (Table 1), and merits further assessment of congruence with other nonmolecular datasets (e.g., morphology). This combined approach should enhance the species-level taxonomy in a genus in which it is extremely difficult to correctly identify species (see Solari, 2007).
The following accounts are provided as a first step toward the definition of the groups recovered in this study. For each species group, a provisional name based on the oldest species name available within each is provided, along with previous references to similar arrangements. Taxonomic composition and geographic distribution are based on specimens studied, but also on information included in earlier works. Departures from current taxonomic usage (Gardner, 2005; Pine and Handley, 2008) are indicated.
The brevicaudata species group
Species in this group occur throughout the tropical dry forests of northern South America, eastern Amazonia from the Guiana Shield to central Brazil, the Cerrado grasslands, and the lowlands of western Amazonia (Fig. 1). Species included in this species group are brevicaudata Erxleben, glirina Wagner, domestica Wagner, plus 'species B' and 'species D' of Pine and Handley (2008); however, because the specimen representing the latter was not recovered as distinct and clustered within brevicaudata, it is no longer considered as distinct. The only indication of a likely affinity among the included species is that many of them were listed as synonyms or subspecies of brevicaudata (e.g., Cabrera, 1958). Similarly, domestica was suggested as close to brevicaudata sensu lato when Pine (1980) compared these forms to maraxina- see also Patton and Costa (2003), Pine and Handley (2008). The following taxa should also be included in this species group: palliolata Osgood and maraxina Thomas (see Pine, 1980; Voss et al., 2001). Individuals allocated to orinoci by Reig et al. (1977), Linares (1998), and Ventura et al. (1998) actually represent an unnamed form referred to as 'species A' by Pine and Handley (2008); however, orinoci is a synonym of brevicaudata (Voss et al., 2001). Node support for this species group was over 90 in both ML and MP, and 1.00 for PP (Table 1).
Gomes (1991) allocated the taxa of this species group to three different groups: touan (including glirina, touan, and 'amazonica' [nomen nudum]), brevicaudata (brevicaudata, rubidus [sic], and 'macae' [nomen nudum]), and domestica. His definitions for these taxa are not comparable to those in use by other authors (Voss et al., 2001; Gardner, 2005; Pine and Handley, 2008), and will not be discussed further. In addition, Gomes (1991) listed all of the Andean species (adusta plus kunsi groups of this analysis) under his brevicaudata. The tricolored and bicolored forms of brevicaudata sensu Voss et al. (2001) are the basis for a restricted use of touan and brevicaudata, respectively. A putative relationship between emiliae and b. touan (proposed by Carvalho, 1960) was discussed and refuted by Pine and Handley (1984).
Relationships among species are consistent in all analyses, with brevicaudata as sister to domestica and to (glirina + species B). Sequence divergence ranges from 8.65% (glirina - species B) to 13.99% (brevicaudata -glirina). Average within divergence ranges from 2.69% (domestica) to 4.84% (brevicaudata). Subclades of M. brevicaudata (6.9% of divergence) correspond to the "tricolor" and "bicolor" forms discussed by Voss et al. (2001) and the phylogroups identified by Steiner and Catzeflis (2004). Subclades of M. glirina are geographically separated by the Beni-Madeira drainage, which is a major barrier in lowland Amazonia (Cracraft, 1985), and diverge by 7.7%. Samples of M. domestica come from Brazil, Bolivia, and Paraguay; minor geographic structure is caused by samples from central Brazil being sister to the clade formed by the other sequences. The single specimen of 'species B' of Pine and Handley (2008) comes from eastern Bolivia, and represents an undescribed species.
The adusta species group
These taxa are associated with the Andean Cordillera, along an elevational range that includes the lowland rainforests to montane forests, with M. reigi being a geographic outlier (Fig. 1; see also Lim et al., 2010) to this pattern. This species group includes adusta Thomas, peruviana Osgood, osgoodi Doutt, and handleyi Solari. This species group was suggested by Cabrera (1958) and Anderson (1997), but defined and restricted by Solari (2007); as delimited here, this group does not include kunsi (see Anderson, 1997). Other taxa, not sampled but considered as part of this group, based on morphological traits, are melanops Goldman, ronaldi Solari, and reigi Lew and Perez-Hernandez. Lim et al. (2010) supported both, the recognition of the adusta group of species (node support was 95 for BS and 1.00 for PP) and the position of reigi within this group. In the present topology, node support was rather low (BS < 70; PP < 0.80; Table 1) in all analyses.
Although this group has generally been recognized by other authors, its integrity was challenged when Gomes (1991) listed some of the abovelisted names under his concept of M. brevicaudata. Morphometric affinities were recognized between adusta and reigi (Ventura et al., 2005), as opposed to members of the brevicaudata group (as defined here), based on Venezuelan specimens only. Morphological resemblance and nearby distribution suggested a relationship between members of the adusta group and kunsi (see Anderson, 1997; Vargas et al., 2003) but analyses of genetic data show these as distantly related lineages.
The internal topology is the same as that presented by Solari (2007) and Lim et al. (2010), with adusta being sister to the remaining species, followed by reigi, and with a sister relationship between peruviana and osgoodi plus handleyi. Genetic divergences among species range from 8.97% for peruviana -osgoodi, to 14.55% for adusta - handleyi. The geographic sub-clades in osgoodi diverge by over 6%, which is similar to the average internal divergence in peruviana (Solari, 2007). Samples of M. adusta come from Ecuador and northern Peru, whereas handleyi come from a single locality in northeastern Peru. Samples of peruviana come from northern through southern Peru and central Bolivia; this species is parapatric with osgoodi at the southern part of its range.
The dimidiata species group
The range of this species group includes very different habitats: the Pampa, the Chaco, and Atlantic forests (Flores, 2006; Pine and Handley, 2008), all located around southeastern South America (Fig. 1). Only two species are included in this group, dimidiata Wagner, and sorex Hensel. These taxa were grouped together by Pine et al. (1985), also including fosteri Thomas and henseli Thomas, the former tentatively regarded as a full species and the latter as a synonym of sorex. It is worth noting that brevicaudis Olfers 1818 (a nomen oblitum according to Pine and Handley, 2008) is sometimes used as the oldest name for sorex or for a distinct species (Gomes, 1991; Brown, 2004 [following Hershkovitz, 1959]). This group has bootstrap support of 100 for MP and ML, and 1.00 for PP (Table 1).
Although sorex is recognized as a distinct species (Pine et al., 1985; Gardner, 2005; Pine and Handley, 2008), the taxa do not form reciprocal monophyletic groups as well as show very low internal divergence (< 0.5%), similar to the range found in among populations of a single species of recent divergence. Miranda et al. (2007) found low levels of divergence among oryzomyine rodents in the transition between Atlantic forests and Pampa of southeastern Brazil; these authors suggested that recent changes molded by ocean level fluctuations may resulted in low geographic structure over this large distribution (despite habitat distinctions). It is possible that dimidiata and sorex (or even fosteri, as suggested by Pine et al., 1985) represent a single species with disjoint geographic ranges and distinct coloration patterns. If that is the case, the oldest available name would be dimidiata Wagner 1847.
The theresa species group
This is the only species group restricted to the Atlantic forests. There are two species included in this group, theresa Thomas and scalops Thomas. A relationship of these names had not been suggested, although both occur in southeastern Brazil. However, Gomes (1991) suggested that dorsal stripes and brownish to reddish dorsal coloration (as seen in theresa) represent a juvenile pelage that change into an adult pattern, with gray on the dorsum, reddish to orangish head, rump and limbs, and no dorsal stripes (as seen in scalops). Recent findings by L. P. Costa and collaborators (pers. comm.) indicate this could be an age dependent trait, thus supporting Gomes (1991) hypothesis. Because the presence of dorsal stripes is recorded in at least other two groups, this state character may have evolved more than once within Monodelphis. Node support is over 90 for the ML and MP analyses, and 1.00 for PP (Table 1).
Average genetic divergence between theresa and scalops is only 3.4%, whereas internal divergence in theresa reaches 2.5%; these values are within the range seen in species with large geographic ranges (e.g., brevicaudata, peruviana). All the theresa specimens come from Sao Paulo, whereas the single scalops comes from Minas Gerais (Fig. 1). If further examination of the animal from Minas Gerais reveals it represents an adult with "typical" scalops coloration, the affinities suggested by Gomes (1991) would be verified.
The emiliae complex
The species is restricted to the lowland forests in a westeast band of Amazonia in Brazil, Bolivia, and Peru (Fig 1). Only one species is included, emiliae Thomas. No other taxon has been associated with emiliae, except by erroneous interpretations (tricolor emiliae in Miranda-Ribeiro, 1936, or touan emiliae in Cabrera, 1958) that made emiliae a subspecies within the brevicaudata group, an issue addressed by Pine and Handley (1984). Linares (1998) also treated this species as M. touan, which is a synonym of M. brevicaudata (see Voss et al., 2001). Resemblance between emiliae and scalops only applies to overall color pattern (see Pine and Abravaya, 1978). Node support is > 95 in both MP and ML bootstrap, and 1.00 for PP (Table 1). Average within divergence is 0.5%, with little geographic structure; although it might be explained by inadequate sampling.
The kunsi complex
The geographic range shown by the samples includes southern Bolivia, northern Argentina, and eastern Paraguay (Fig. 1), which, when combined with verified records from northern Bolivia and central Brazil (see De la Sancha et al., 2007), makes it rather biogeographically complex. This is another species group with only one species, kunsi Pine, which has sometimes been associated with adusta and related taxa (Anderson, 1997), but the latter form a distant cluster herein recognized as a species group. Although kunsi and some members of the adusta group share a uniform buffy to light-brownish overall coloration, fur color seems to be homoplastic as it is seen in other taxa (e.g., red sides in brevicaudata and sorex, or black dorsal stripes in theresa and americana). Node support is 100 for MP and ML trees, and 1.00 for PP (Table 1).
The average internal divergence is 0.8%, compatible with the fact that only one species is represented; and, if some geographic structure can be recovered, it would account for only 1.5% between Bolivia-Argentina and Paraguay. A recent comparison with GenBank sequences from central Brazil assigned to kunsi (submitted by B. A. Carvalho et al.) clusters these as sister to the group recovered in this study with almost 6.5% divergence, which is a high value but below the average among recognized species in this study (above 8.5% in the adusta or brevicaudata groups). This may indicate a biogeographic division between those populations.
The americana complex
For this species, Pine and Handley (2008) documented an extensive range in eastern Brazil. Although only one species, americana Muller, forms this group, more taxa may be included. Currently, this species name is used for the three-dorsal-striped taxon from eastern Brazil, extending from north to south (as opposed to others restricted to the Atlantic forest, such as iheringi or theresa). Node support is > 90 in the MP and ML trees, and 1.00 for PP (Table 1). Voss and Jansa (2003) suggested that an americana species group could be associated with other dorsally striped animals (such as theresa), but these taxa did not fall together in the present phylogenetic analyses. Samples from two localities, Bahia and São Paulo (Fig. 1), clustered together with 8.1% of divergence, which is larger than average for other species in this study and could then represent a subspecific division. In Gomes's (1991) view, americana includes a northern and a southern group which represent two different species, but provided no names for these.
The 'species C' complex
The only species in this group has not been named. So far, the species is known from four montane forests at elevations above 1800 m in central Peru (Solari et al., in prep.). Although this form was called theresa by Gardner (1993), Pacheco et al. (1995), and Patton and Costa (2003), no relationship with that species has been found. Node support is 100 in MP and ML trees, and 1.00 for PP (Table 1). The present analysis verified that distinction at a genetic level; in fact, although the 'species C' of Pine and Handley (2008) is remarkably similar to americana, these two exhibit a high genetic divergence (> 15%) from each other. Samples from two localities in central Peru (Fig. 1) diverge by 3.5%; similar geographic divergence was found when compared with shorter sequences (~400 bp) from other localities. Although the unnamed species appeared as sister to americana in the Bayesian topology, this relationship had low support (< 0.65) so they are recognized as independent lineages in this arrangement.
TAXONOMIC CONCLUSIONS
Based on these results, it is clear that most of the species groups recognized by some authors (e.g., Matschie, 1916; Cabrera, 1919; Gilmore, 1941; Gomes, 1991) do not correspond to monophyletic groups. Therefore, a taxonomic arrangement consistent with the hypotheses of relationship for the included taxa is presented and discussed in the following section.
The oldest available name for the genus of shorttailed opossums is Monodelphis Burnett 1830, with Monodelphis brachyura: Burnett (=Didelphis brevicaudata Erxleben 1777) as type species. Another genus name, Hemiurus Gervais 1855, was coined to include brachyurus, tristriatus, and tricolor, but the name is preoccupied by Hemiurus Rudolphi 1809 (Platyhelminthes) and therefore is unavailable (see Pine and Handley, 2008).
Peramys Lesson 1842 was proposed with no type species; later Thomas (1888) designated P. brachyura: Lesson (=M. dimidiata, see Pine and Handley, 2008) as type species. Cabrera (1919) proposed Minuania as a subgenus with Didelphys dimidiata Wagner 1847 as type species, and it is therefore a subjective junior synonym of Peramys. Then, Miranda-Ribeiro (1936) described and included dimidiata itatiayae and umbristriata (goyana of the same author is a synonym), and Cabrera and Yepes (1940) referred fosteri to Minuania. Haltenorth (1958) listed Minuania as full genus including dimidiata, fosteri, and umbristriata, whereas Pine (1976) treated it as a valid subgenus with dimidiata and umbristriata only. Kirsch and Calaby (1977) did not recognize Minuania as separate from Monodelphis, and listed dimidiata (with fosteri as a synonym) but not umbristriata. Pine et al. (1985) tentatively treated henseli Thomas as a synonym of sorex, and fosteri as a full species. Another genus-group name is Monodelphiops Matschie 1916, with Microdelphys sorex Hensel 1872 as type species; however, because of the close relationships between M. dimidiata and M. sorex, Monodelphiops Matschie, with M. sorex as type species, should be a synonym of Peramys.
Microdelphys Burmeister 1856 was proposed as a subgenus of Didelphis, with tristriata: Burmeister (=Sorex americanus Müller 1776) as type species by subsequent designation (Thomas 1888). Miranda-Ribeiro (1936) listed americana, iheringi, theresa, and unistriata in a key for subgenus Microdelphys; however, this arrangement conflicts with results presented here by inclusion of theresa. In a restricted sense, Microdelphys would include short-tailed opossums with blackish dorsal stripes that are conspecific with or that share a close relationship with the type species (M. americana).
Gilmore's (1941) use of names Monodelphis ("tricolor" group), Lestodelphys ("bicolor" group), Microdelphys ("striped" group), and Minuania ("variegated" group), as subgenera of Monodelphis is difficult to evaluate because no species were listed under them. In any case, his definition of Monodelphis is polyphyletic by inclusion of Lestodelphys. Dismissing Gilmore's system, the arrangement for the five available genus-group names would stand as follows:
1. Monodelphis Burnett 1830 (Type species: brachyura: Burnett = brevicaudata Erxleben)
2. Peramys Lesson 1842 (Type species: brachyurus: Lesson = dimidiata Wagner)
Monodelphiops Matschie 1916 (Type species: sorex Hensel)
Minuania Cabrera 1919 (Type species: dimidiata Wagner)
3. Microdelphys Burmeister 1856 (Type species: tristriata: Burmeister = americana Muller).
Strict application of the resulting phylogenies to update the subgeneric arrangement would result in a restricted meaning for the three genus-group names. Thus, Monodelphis would include the brevicaudata group as defined above, and finally, Microdelphys should be restricted to the americana group, as defined above, and excluding the other species groups (M. theresa and 'species C') displaying dorsal stripes. Four recognized species that were not included in the phylogenetic analyses (iheringi, rubida, umbristriata, unistriata) would remain as incertae sedis within Monodelphis (sensu lato).
Given the low resolution of the topologies in regard to relationships among the eight species groups, Monodelphis (sensu stricto), Peramys, Microdelphys, and the remaining five (still unnamed), they should be considered of similar rank and not to be arranged into more inclusive groups. Under a conservative approach, Monodelphis (sensu lato) would stand as the valid name for the genus and these eight species groups could be regarded as equivalent to subgenera but without use of formal taxon names. Use of a single genus name seems to be the best choice to promote stability, and this still allows for recognition of the group's extensive supraspecific diversity through explicit treatment of its contained species groups. Use of a different genetic marker, one with a slower rate of substitution (such as a nuclear intron; e.g., DMP1, see Jansa et al., 2006), for the same set of taxa studied here should provide a powerful starting point to assess the constraints and informative value of the cytochrome b gene for Monodelphis. This, along with an improved taxon sampling, would be necessary to resolve definitions and limits for Monodelphis and its included species.
List of specimens examined including species name, geographic locality, GenBank accession and voucher numbers for 60 Monodelphis and six outgroup cytochrome-b sequences. Asterisks (*) by tissue numbers denote 25 sequences (20 for ingroup and five for outgroup) obtained from GenBank. Sequences by J. L. Patton and collaborators are indicated by a karat (^). Museum catalog numbers (see 'Material and Methods' for acronyms) are missing for vouchers housed but not yet cataloged or for which the number is unknown. Numbers in parentheses following the localities make reference to symbols used in Fig. 1. Ma. = Marmosa, M. = Monodelphis; JPJ = J. P. Jayat (Laboratorio de Investigaciones Ecológicas de las Yungas, Universidad Nacional de Tucumán, Argentina); JAA = J. Amanzo A. (MUSM, Lima, Peru).

ACKNOWLEDGMENTS
A critical set of sequences used in these analyses was obtained by J. L. Patton and L. P. Costa (Museum of Vertebrate Zoology, University of California, Berkeley); their support is greatly appreciated. S. R. Hoofer (Texas Tech University, now at the University of Kansas, Lawrence) designed specific primers used in this and other studies, and greatly helped in several steps of these analyses. W. Munera (U de Antioquia) helped with preparation of the map for Fig. 1. For loan of tissues, thanks are extended to T. Tarifa and J. Vargas of the Colección Boliviana de Fauna (La Paz, Bolivia); J. P. Jayat of the Laboratorio de Investigaciones Ecológicas de las Yungas (Tucumán, Argentina); B. D. Patterson and W. Stanley of the Field Museum (Chicago), E. Lessa and G. D'Elía (now at Universidad de Concepción, Chile) of the Laboratorio de Evolución, Universidad de la República (Montevideo, Uruguay); V. Pacheco, C. Tello, and S. Velazco of the Museo de Historia Natural, Universidad Nacional Mayor de San Marcos (Lima, Perú); T. L. Yates and S. Parmenter of the Museum of Southwestern Biology, University of New Mexico (Albuquerque); J. L. Patton and C. Cicero of the Museum of Vertebrate Zoology, University of California (Berkley); and R. J. Baker and H. Garner of the Natural Science Research Laboratory (NSRL), Texas Tech University (Lubbock). R. H. Pine (University of Kansas, Lawrence) and two anonymous reviewers made suggestions concerning the manuscript. Labmates at Baker's Lab and the NSRL provided valuable assistance during my research at Texas Tech. Support from Texas Tech University through a Chancellor SBC Fellowship (2002-2005), a J Knox Jones Memorial Fund (2004), and a Cash Family Endowment (2006) scholarship was also provided. I was also supported by an Albert R. and Alma Shadle Fellowship (2005) and an ASM Fellowship (2006), from the American Society of Mammalogists. Additional support received from Texas Tech University through a line item for biodiversity studies to R. J. Baker.
LITERATURE CITED
1. ANDERSON S. 1997. Mammals of Bolivia. Taxonomy and distribution. Bulletin of the American Museum of Natural History 231:1-652. [ Links ]
2. AVISE JC. 2004. Molecular markers, natural history, and evolution. Second Edition. Sinauer Associates Inc., Sunderland, Massachusetts. [ Links ]
3. BAKER RJ and RD BRADLEY. 2006. Speciation in mammals and the genetic species concept. Journal of Mammalogy 87:643-662. [ Links ]
4. BROWN BE. 2004. Atlas of New World marsupials. Fieldiana: Zoology 102:1-308. [ Links ]
5. CABRERA A. 1919. Genera Mammalium. Monotremata, Marsupialia. Museo Nacional de Ciencias Naturales, Madrid, Spain, 177 pp., 19 pl. [ Links ]
6. CABRERA A. 1958. Catálogo de los mamíferos de América del Sur, I. Revista del Museo Argentino de Ciencias Naturales "Bernardino Rivadavia," Ciencias Zoológicas 4:1-307. [ Links ]
7. CABRERA A and J YEPES. 1940. Mamíferos Sudamericanos (vida, costumbres y descripción). Historia Natural Ediar. Compañía Argentina de Editores, Buenos Aires, Argentina. [ Links ]
8. CARVALHO B de A, LFB OLIVEIRA, AP NUNES, and MS MATTEVI. 2002. Karyotypes of nineteen marsupial species from Brazil. Journal of Mammalogy 83:58-70. [ Links ]
9. CARVALHO CT de. 1960. Sobre alguns mamiferos do sudeste do Pará. Arquivos de Zoologia do Estado de São Paulo 11:121-132. [ Links ]
10. COSTA LP. 2003. The historical bridge between the Amazon and Atlantic Forest of Brazil: A study of molecular phylogeny with small mammals. Journal of Biogeography 30:71-86. [ Links ]
11. COSTA LP, YL LEITE, and JL PATTON. 2003. Phylogeography and systematic notes on two species of gracile mouse opossums, genus Gracilinanus (Marsupialia: Didelphidae) from Brazil. Proceedings of the Biological Society of Washington 116:275-292. [ Links ]
12. CRACRAFT J. 1985. Historical biogeography and patterns of diversification within South American areas of endemism. Pp. 49-84, in: Neotropical Ornithology (PA Buckley, MS Foster, ES Morton, RS Ridgely, and FG Buckley, eds.). American Ornithological Union, Washington, D.C. [ Links ]
13. CREIGHTON GK. 1984. Systematic studies on opossums (Didelphidae) and rodents (Cricetidae). Ph.D. dissertation, University of Michigan, Ann Arbor. [ Links ]
14. De la SANCHA N, S SOLARI, and RD OWEN. 2007. First records of Monodelphis kunsi Pine (Didelphimorphia: Didelphidae) from Paraguay, with an evaluation of its distribution. Mastozoología Neotropical 14:241-247. [ Links ]
15. FELSENSTEIN J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39:783-791. [ Links ]
16. FLORES DA. 2006. Orden Didelphimorphia. Pp. 31-45, in: Mamíferos de Argentina: sistemática y distribución (RM Barquez, MM Díaz, and RA Ojeda, eds.). Sociedad Argentina para el Estudio de los Mamíferos, Tucumán, Argentina. [ Links ]
17. GARDNER, AL. 1993. Order Didelphimorphia. Pp. 3-18, in: Mammal species of the world. A taxonomic and geographic reference (DE Wilson and DM Reeder, eds.). Second Edition. Smithsonian Institution Press, Washington, D.C. [ Links ]
18. GARDNER AL. 2005. Order Didelphimorphia. Pp. 15-23, in: Mammal species of the world. A taxonomic and geographic reference (DE Wilson and DM Reeder, eds.). Third Edition. The Johns Hopkins University Press, Baltimore, Maryland. [ Links ]
19. GILMORE RM 1941. Zoology. Pp. 314-319, in: The susceptibility to yellow fever of the vertebrates of eastern Colombia. I. Marsupialia (JC Bugher, J Boshell-Manrique, M Roca-Garcia, and RM Gilmore, eds.). American Journal of Tropical Medicine 21:309-333. [ Links ]
20. GOIN FJ and P REY. 1997. Sobre las afinidades de Monodelphis Burnett, 1830 (Mammalia: Marsupialia: Didelphidae: Marmosinae). Neotrópica 43:93-98. [ Links ]
21. GOMES NF. 1991. Revisão sistemática do gênero Monodelphis (Marsupialia). M. S. thesis, Universidade de Sao Paulo, Brazil, 181 pp. [ Links ]
22. HALTERNORTH T. 1958. Die Klassifikation der Saugetiere: Monotremata und Marsupialia. Handbuch der Zoologie, Band 8, Lieferung 16:1-40. [ Links ]
23. HERSHKOVITZ P. 1959. Nomenclature and taxonomy of the Neotropical mammals described by Olfers, 1818. Journal of Mammalogy 40:337-353. [ Links ]
24. HUELSENBECK JP and F RONQUIST. 2001. MrBayes: Bayesian inference of phylogeny. Bioinformatics 17:754-755. [ Links ]
25. JANSA SA and RS VOSS. 2000. Phylogenetic studies on didelphid marsupials. I. Introduction and preliminary results from nuclear IRBP gene sequences. Journal of Mammalian Evolution 7:43-77. [ Links ]
26. JANSA SA, JF FORSMAN, and RS VOSS. 2006. Different patterns of selection on the nuclear genes IRBP and DMP-1 affect the efficiency but not the outcome of phylogeny estimation for didelphid marsupials. Molecular Phylogenetics and Evolution 38:363-380. [ Links ]
27. JANSA SA and RS VOSS. 2005. Phylogenetic relationships of the marsupial genus Hyladelphys based on nuclear gene sequences and morphology. Journal of Mammalogy 86:853-865. [ Links ]
28. KIRSCH JAW and JH CALABY. 1977. The species of living marsupials: An annotated list. Pp. 9-26, in: The biology of marsupials (B Stonehouse and D Gilmore, eds.). University Park Press, Baltimore, Maryland. [ Links ]
29. LANGGUTH A and JFS LIMA. 1988. The karyotype of Monodelphis americana (Marsupialia - Didelphidae). Revista Nordestina de Biologia 6:1-5. [ Links ]
30. LIM BK, MD ENGSTROM, JC PATTON, and JW BICKHAM. 2010. Molecular phylogenetics of Reig's short tailed opossum (Monodelphis reigi) and its distributional range extension into Guyana. Mammalian Biology 75:287-293. [ Links ]
31. LINARES OJ. 1998. Mamíferos de Venezuela. Sociedad Conservacionista Audubon de Venezuela, Caracas, Venezuela. [ Links ]
32. LONGMIRE JL, M MALTBIE, and RJ BAKER. 1997. Use of "lysis buffer" in DNA isolation and its implications for museum collections. Occasional Papers, Museum of Texas Tech University 163:1-3. [ Links ]
33. MATSCHIE P. 1916. Bemerkungen über die Gattung Didelphis L. Sitzunsberichte der Gesellschaft Naturforschender Freunde zu Berlin 1916:259-272. [ Links ]
34. MIRANDA GB, J ANDRADES-MIRANDA, LFB OLIVEIRA, A LANGGUTH, and MS MATTEVI. 2007. Geographic patterns of genetic variation and conservation consequences in three South American rodents. Biochemical Genetics 45:839-856. [ Links ]
35. MIRANDA-RIBEIRO A de. 1936. Didelphia ou Mammalia-Ovovivipara. Marsupiaes, didelphos, pedimanos ou metatherios. Revista do Museu Paulista 20:245-424. [ Links ]
36. MUSTRANGI MA and JL PATTON. 1997. Phylogeography and systematics of the slender mouse opossum Marmosops (Marsupialia, Didelphidae). University of California, Publications in Zoology 130:1-86. [ Links ]
37. PACHECO V, H DE MACEDO, E VIVAR, CF ASCORRA, R ARANACARDO, and S SOLARI. 1995. Lista anotada de los mamíferos peruanos. Occasional Papers in Conservation Biology 2:1-35. [ Links ]
38. PALMA RE and TL YATES. 1996. The chromosomes of Bolivian didelphid marsupials. Occasional Papers, Museum of Texas Tech University 162:1-20. [ Links ]
39. PATTON JL and LP COSTA. 2003. Molecular phylogeography and species limits in rainforest didelphid marsupials of South America. Pp. 63-81, in: Predators with pouches: The biology of carnivorous marsupials (M Jones, C Dickman, and M Archer, eds.). CSIRO Publishers, Collingwood, Australia. [ Links ]
40. PATTON JL and MNF DA SILVA. 1997. Definition of species of pouched four-eyed opossums (Didelphidae, Philander). Journal of Mammalogy 78:90-102. [ Links ]
41. PATTON JL, SF DOS REIS, and MNF DA SILVA. 1996. Relationships among didelphid marsupials based on sequence variation in the mitochondrial cytochrome b gene. Journal of Mammalian Evolution 3:3-29. [ Links ]
42. PATTON JL, MNF DA SILVA, and JR MALCOLM. 2000. Mammals of the Rio Juruá and the evolutionary and ecological diversification of Amazonia. Bulletin of the American Museum of Natural History 244:1-306. [ Links ]
43. PINE RH. 1976. Monodelphis umbristriata (A. de Miranda-Ribeiro) is a distinct species of opossum. Journal of Mammalogy 57:785-787. [ Links ]
44. PINE RH. 1980. Taxonomic notes on "Monodelphis dimidiata itatiayae (Miranda-Ribeiro)", Monodelphis domestica (Wagner) and Monodelphis maraxina Thomas (Mammalia: Marsupialia: Didelphidae). Mammalia 43:495-499. [ Links ]
45. PINE RH and JP ABRAVAYA. 1978. Notes on the Brazilian opossum Monodelphis scalops (Thomas) (Mammalia: Marsupialia: Didelphidae). Mammalia 42:379-382. [ Links ]
46.PINE RH and CO HANDLEY, JR. 1984. A review of the Amazonian short-tailed opossum Monodelphis emiliae (Thomas). Mammalia 48:239-245. [ Links ]
47. PINE RH and CO HANDLEY, JR. 2008. Genus Monodelphis Burnett 1830. Pp. 82-107, in: Mammals of South America, Vol. 1: Marsupials, xenarthrans, shrews, and bats (AL Gardner, ed.). University of Chicago Press, Chicago, Illinois. [ Links ]
48. PINE RH, PL DALBY, and JO MATSON. 1985. Ecology, postnatal development, morphometrics, and taxonomic status of the short-tailed opossum, Monodelphis dimidiata, an apparently semelparous annual marsupial. Annals of the Carnegie Museum 54:195-231. [ Links ]
49. POSADA D and KA CRANDALL. 1998. Modeltest: Testing the model of DNA substitution. Bioinformatics 14:817-818. [ Links ]
50. REIGOA, AL GARDNER, NO BIANCHI, and JL PATTON. 1977. The chromosomes of the Didelphidae (Marsupialia) and their evolutionary significance. Biological Journal of the Linnean Society, London 9:191-216. [ Links ]
51. REIG OA, JAW KIRSCH, and LG MARSHALL. 1987. Systematic relationships of the living and Neocenozoic American opossumlike marsupials (suborder Didephimorphia), with comments on the classification of these and of the Cretaceous and Paleogene New World and European metatherians. Pp. 1-90, in: Possums and opossums: Studies in evolution (M Archer, ed.). Surrey Beatty and Sons Pty. Ltd. and Royal Zoological Society of New South Wales, Sydney, Australia. [ Links ]
52. SAMBROOK J, EF FRITSCH, and T MANIATIS. 1989. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York. [ Links ]
53. SOLARI S. 2004. A new species of Monodelphis (Didelphimorphia: Didelphidae) from southeastern Peru. Mammalian Biology 69:145-152. [ Links ]
54. SOLARI S. 2007. New species of Monodelphis (Didelphimorphia: Didelphidae) from Peru, with notes on M. adusta (Thomas 1897). Journal of Mammalogy 88:319-329. [ Links ]
55. SPRINGER MS, RW DEBRY, C DOUADY, HM AMRINE, O MADSEN, WW DE JONG, and MJ STANHOPE. 2001. Mitochondrial versus nuclear gene sequences in deeplevel mammalian phylogeny reconstruction. Molecular Biology and Evolution 18:132-143. [ Links ]
56. STEINER C and FM CATZEFLIS. 2004. Genetic variation and geographical structure of five mouse-sized opossums (Marsupialia, Didelphidae) throughout the Guiana region. Journal of Biogeography 31:959-973. [ Links ]
57. STEINER C, M TILAK, EJP DOUZERY, and FM CATZEFLIS. 2005. New DNA data from a transthyretin nuclear intron suggest an Oligocene to Miocene diversification of living South America opossums (Marsupialia: Didelphidae). Molecular Phylogenetics and Evolution 35:363-379. [ Links ]
58. SWOFFORD DL. 2002. PAUP*: Phylogenetic analysis using parsimony (*and other methods), version 4.0 b10. Sinauer Associates, Sunderland, Maryland. [ Links ]
59. THOMAS O. 1888. Catalogue of the Marsupialia and Monotremata in the collection of the British Museum (Natural History). Trustees of the British Museum (Natural History), London. xiv + 401 pp. [ Links ]
60. VARGAS J, T TARIFA, and C CORTEZ. 2003. Nuevos registros de Monodelphis adusta y Monodelphis kunsi (Didelphimorphia: Didelphidae) para Bolivia. Mastozoología Neotropical 10:123-131. [ Links ]
61. VENTURA J, R PEREZ-HERNANDEZ, and MJ LOPEZ-FUSTER. 1998. Morphometric assessment of the Monodelphis brevicaudata group (Didelphimorphia: Didelphidae) in Venezuela. Journal of Mammalogy 79:104-117. [ Links ]
62. VENTURA J, D LEW, R PEREZ-HERNANDEZ, and MJ LOPEZ-FUSTER. 2005. Skull size and shape relationships between Venezuelan Monodelphis taxa (Didelphimorphia: Didelphidae), including the recently described species M. reigi Lew & Pérez-Hernández. Tropical Zoology 18:227-235. [ Links ]
63. VOSS RS and S JANSA. 2003. Phylogenetic studies on didelphid marsupials II. Nonmolecular data and new IRBP sequences: Separate and combined analyses of didelphine relationships with denser taxon sampling. Bulletin of the American Museum of Natural History 276:1-82. [ Links ]
64. VOSS RS and S JANSA. 2009. Classification of Didelphid marsupials, an extant radiation of New World metatherian mammals. Bulletin of the American Museum of Natural History 322:1-178. [ Links ]
65. VOSS RS, DP LUNDE, and NB SIMMONS. 2001. The mammals of Paracou, French Guiana:A Neotropical lowland rainforest fauna. Part 2. Nonvolant species. Bulletin of the American Museum of Natural History 263:1-236. [ Links ]














