Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
Mastozoología neotropical
versión impresa ISSN 0327-9383
Mastozool. neotrop. vol.21 no.1 Mendoza jun. 2014
ARTÍCULO
Chromosomal variability and morphological notes in Graomys griseoflavus (Rodentia, Cricetidae, Sigmodontinae), from Catamarca and Mendoza provinces, Argentina
Cecilia Lanzone1, 2, Schullca N. Suárez1, Daniela Rodriguez1, Agustina Ojeda1, Soledad Albanese1, and Ricardo A. Ojeda1
1 Grupo de Investigaciones de la Biodiversidad (GiB), CONICET-CCT-Mendoza-IADIZA, Mendoza, Argentina [correspondence: Cecilia Lanzone <celanzone@mendoza-conicet.gob.ar>].
2 Laboratorio de Genética Evolutiva, FCEQyN, IBS, UNaM-CONICET, Félix de Azara 1552, CPA N3300LQF Posadas, Misiones, Argentina.
ABSTRACT.
Genetic variability in rodents is extremely wide and a fruitful field of research. Graomys griseoflavus is a phyllotine rodent, endemic to South America, polymorphic for Rb rearrangements. However, few individuals and populations were studied cytogenetically to date, considering its wide distribution. We present and compare chromosomal data from Mendoza and Catamarca provinces, contrasting previous hypothesis about its karyotypic evolution. All populations were polymorphic for Rb rearrangements; in addition, we describe a new fusion from Mendoza. The presence of more than one heterozygous fusion in several localities refute the hypothesis proposed for this species that for a new fusion to be generated the others must occur in homozygosis. The recorded 2n have an irregular geographic distribution. The extra short arms detected are additional factors of chromosome variability. Some external qualitative characters (i.e., coloration) show certain variability. In some quantitative external and cranial characters, a low degree of sexual dimorphism was detected. However, there were not significant differences in external and cranial metrics variables among localities indicating low degree of differentiation, as reported in previous works; neither the coefficients of variation of these variables had high values compared to other related species. While a larger sample is needed for these different types of characters, the high chromosomal variability does not seem to correspond with comparable degrees of morphological and mitochondrial variability in G. griseoflavus.
RESUMEN.
Variabilidad cromosómica y notas morfológicas en Graomys griseoflavus (Rodentia, Cricetidae, Sigmodontinae) de las provincias de Catamarca y Mendoza, Argentina.
La variabilidad genética de los roedores es extremadamente amplia y es un fructuoso campo de investigaciones. Graomys griseoflavus es un roedor filotino endémico de América del Sur, polimórfico para reordenamientos Rb. Si bien posee una extensa distribución, pocos individuos y poblaciones han sido estudiados citogenéticamente. En este trabajo presentamos y comparamos datos cromosómicos de las provincias de Mendoza y Catamarca, contrastando hipótesis previas sobre su evolución cariotípica. Todas las poblaciones fueron polimórficas para rearreglos Rb y se describe una nueva fusión en ejemplares de Mendoza. La presencia de más de una fusión en heterocigosis en varias localidades refuta la hipótesis propuesta para esta especie de que para la generación de una nueva fusión las otras deben presentarse primero en homocigosis. Los citotipos registrados poseen una distribución geográfica irregular. La detección de brazos cortos extras muestra factores adicionales de variabilidad cromosómica. Algunos caracteres cualitativos externos (i.e., coloración) muestran cierta variabilidad. En algunos caracteres cuantitativos externos y del cráneo se detectó un bajo grado de dimorfismo sexual. Sin embargo, no hubo diferencias significativas entre localidades en las variables métricas externas y craneales, como se reporta en otros trabajos para la especie; tampoco los coeficientes de variación de esas variables presentaron valores elevados comparados con otras especies relacionadas. Si bien una muestra mayor es necesaria para estos diferentes tipos de caracteres, la alta variabilidad cromosómica parece no ser correspondida con grados comparables de variabilidad morfológica y mitocondrial en G. griseoflavus.
Key words: Chromosomal rearrangements; Phyllotini; Sigmodontinae.
Palabras clave: Phyllotini; Rearreglos cromosómicos; Sigmodontinae.
Recibido 23 agosto 2013.
Aceptado 30 diciembre 2013.
Editor asociado: P Teta
INTRODUCTION
Sigmodontines are a group of rodents mostly distributed in South America that have high genetic diversity (Smith and Patton, 1993, 1999; Spotorno et al., 2001; D'Elía, 2003; Steppan et al., 2004; Lanzone et al., 2011). Among them, the genus Graomys is a phyllotine found in arid to semiarid regions of the southern portion of South America (Hershkovitz, 1962; Reig, 1986). Several species have been described for Graomys. However, the validity and distribution of some of these forms is confusing (e.g., Graomys edithae is only known from early collections at its type locality). Associated to that, the evolutionary processes that gave rise to the species in the genus are topics of intense debate, a common problem for several other sigmodontine rodents (Hershkovitz, 1962; Tiranti, 1998; Gómez-Laverde et al., 2004; Wilson and Reeder, 2005; Lanzone et al., 2007, 2011).
In Graomys, the junction of chromosome and geographic data allowed the identification of three species with essentially different chromosome complements and distributional ranges. G. griseoflavus presents a variation in chromosome number of 2n = 34 to 2n = 38 due to Robertsonian (Rb) translocations, and in fundamental number (FN) due to inversions. This species is found mostly in the Monte biome (Tiranti, 1998; Theiler and Blanco 1996a; Martínez and Di Cola, 2011). Graomys chacoensis (including Graomys centralis) with 2n = 42 presents inversions that modified its FN and is distributed in the Chaco and Espinal biomes (Zambelli et al., 1994; Tiranti, 1998; Lanzone et al., 2007; Ferro and Martínez, 2009; Martínez and Di Cola, 2011). Finally, Graomys domorum with 2n = 28 is found in the forest and transitional areas of Bolivia and northwestern Argentinean Andes (Hershkovitz, 1962; Pearson and Patton, 1976; Martínez and Di Cola, 2011).
Graomys griseoflavus and G. chacoensis are morphologically undistinguishable species in the field, because there are no simple characters permitting unequivocal identification. Traditional and geometric morphometrics indicated differentiation in size and shape of skull among the 3 species (Ferro and Martínez, 2009; Martínez et al., 2010a; Martínez and Di Cola, 2011), being G. domorum the most divergent of all three species recognized in the genus (Martínez and Di Cola, 2011).
Reproductive and molecular phylogenetic and phylogeographic studies confirmed the specific status of G. griseoflavus and G. chacoensis, which are sister species of recent origin (Zambelli et al., 1994; Zambelli and Vidal-Rioja, 1995; Theiler and Blanco, 1996a, 1996b; Zambelli and Vidal-Rioja, 1999; Ferro and Martínez, 2009; Martínez et al., 2010b). Analyses based on mitochondrial sequences of cytochrome b and control region showed the existence of two distinct clades which clearly correspond with the 2n = 42 cytotype and the Rb complex with 2n = 34-38, respectively (Catanesi et al., 2002, 2006; Martínez et al., 2010b). However, molecular differentiation and variability within and between these clades is relatively scarce in mtDNA D-loop region (Martínez et al., 2010b) —similar to others phyllotine species in mtDNA cytochrome b (Catanesi et al., 2002; Ferro and Martinez, 2009)— and high in levels of allozymic variation and differentiation of some populations (Theiler and Gardenal, 1994; Theiler et al., 1999a).
The evidence indicates that the different cytotypes of G. griseoflavus originated from 2n = 42 cytotypes, and this is consistent with studies on distribution of families of highly repetitive DNA sequences, and molecular cytogenetics of presence and location patterns of nucleolar organizer regions (NOR) in G. griseoflavus (Zambelli and Vidal-Rioja, 1995, 1996, 1999).
It has been suggested that reproductive isolation between these two lineages has been acquired through chromosomal speciation, which would have been caused by Rb fusions (Zambelli et al., 1994; Theiler and Blanco, 1996a). There are two hypotheses about the origin of the Rb variability found in the genus. Both agree that the ancestral species would be G. chacoensis, but disagree on the sequences of events that led to G. griseoflavus. One explained the Rb fusions by the appearance and subsequent fixation of these chromosomal mutations independently and simultaneously on several individuals, without drastic demographic reductions in the founder population (Theiler et al., 1999a). On the other hand, Zambelli et al. (2003) argue a single sequential origin of Rb rearrangements in Graomys from a few individuals of the ancestral population, generated by a founding event that would have resulted in establishment of different chromosomal variations. Until now, none of these two hypotheses can be completely contrasted, in part because few populations and individuals have been studied cytogenetically to date, considering the wide geographic range it has.
In order to extend the study of this polymorphism in G. griseoflavus, we describe the chromosome constitution of individuals from new localities and contrast our results with the proposed hypothesis for chromosome evolution in the species. Additionally, quantitative and qualitative morphological data are presented to investigate in a preliminary way if external and cranial characters have a comparable degree of variability contrasted with chromosome or molecular data.
MATERIAL AND METHODS
We analyzed the chromosomal characteristics of 30 individuals and some quantitative and qualitative characters of 48 specimens from Catamarca and Mendoza provinces, Argentina (Tables 1, 2, and 3 [supplementary material] and Appendix). Chromosome preparations of specimens were obtained using the standard hypotonic technique for bone-marrow (Ford and Hamerton, 1956) with small modifications (Table 1). Chromosomes were stained with Giemsa (pH=6.8). Ten metaphase spreads were counted for each specimen. Fundamental Number of chromosome arms (FN) was determined according to Patton (1967). Chromosomal measurements were taken in selected cells from individuals with different diploid numbers. Only one individual was used for each diploid number, with the exception of those with the new fusion, which were grouped. The absolute values of the chromosome pairs were transformed to percentages of the haploid set. In each category chromosomal measurements were averaged and the means (X) and standard deviation (SD) are reported (Table 2). Also G-banding (Seabright, 1971) and studies of male meiosis (Evans et al., 1964) in some individuals were performed.
Table 1 Localities (see Fig. 1 and Appendix for additional details), coordinates, number of individuals (N), chromosome number (2N), fundamental number (FN), and sex (M = male, F = female) of specimens of Graomys griseoflavus studied in this work.

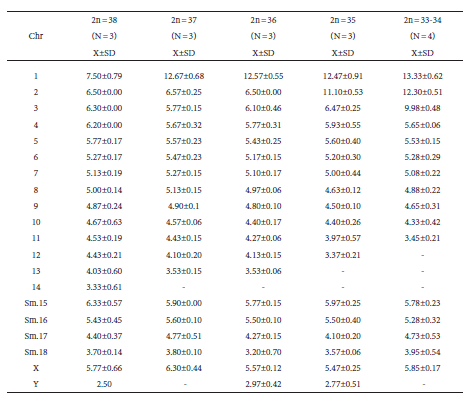
Table 2 Average of chromosomal measurements (as percentage of haploid set), taken on 16 cells with diploid numbers varying from 2n = 33 to 2n = 38, pertaining to different Graomys griseoflavus individuals from localities of Mendoza and Catamarca. N = number of cells used per diploid number; Chr = chromosomal pairs; 2n = diploid number; Sm = submetacentric; X = mean; SD = standard deviation. The specimen with 2n = 38 is from Reserva de Ñacuñán (Mendoza), that with 2n = 37 from Finca La Betania, (Mendoza), the one with 2n = 36 from Cuenca del Maure (Mendoza), and the specimen with 2n = 35 is from Salar de Pipanaco (Catamarca). Specimens with 2n = 33 and 34 are from 30 km S de Mendoza Capital (Mendoza) and were grouped to share the same chromosomes Rb. In heterozygous Rb was measured only the fused chromosome in the trio.
For univariate morphometric analyses, 6 external and 22 standard cranial features were measured (Table 3, supplementary material) using a caliper rounded to the nearest 0.1 mm. (Sikes et al., 1997; Martin et al., 2001). Means, standard deviation (SD), and coefficient of variation (CV) were estimated. Comparisons between sexes were performed with Mann-Whitney U-test. For comparisons among populations, a Kruskal-Wallis test was used. Levels of statistical significance (p) are reported when the differences were significant. Analyses were implemented using Statistica Software. Additional external observations related to dorsal, lateral and ventral coloration are described in the text. Only animals with a completely erupted dentition were used for all analyses and observations.
RESULTS
Karyotypes
All populations were polymorphic for Rb rearrangements (Table 1, Fig. 1). Diploid numbers varied from 2n = 33 to 38, and fundamental numbers from 44 to 45. The karyotypes with 2n = 38 (Fig. 2a) have 14 pairs of acrocentric chromosomes that gradually decrease in size (pairs 1-14), and 4 submetacentric medium and small sized chromosomes (pairs 15, 16, 17 and 18) that are constant among all cytotypes of this species. Individuals with 2n = 37 have one extra metacentric chromosome of large size and are heterozygous for an Rb translocation. Individuals with 2n = 35 have 2 fusions of similar size, 1 in homozygous and the other in heterozygous state (Fig. 2b). Individuals with 2n = 36 can only have 1 Rb homozygous fusion or both fusions in heterozygosis. Individuals with 2n = 33 and with 2n = 34 have extra fusions involving acrocentrics of different size (Fig. 2c).

Fig. 1. Map of Central Argentina showing localities from which Graomys griseoflavus were analyzed in this study; for the reference number see Table 1.
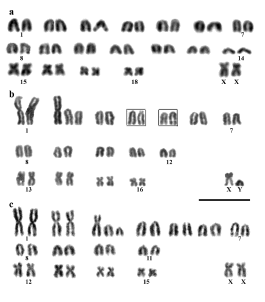
Fig. 2. Bone marrow Giemsa staining karyotype of Graomys griseoflavus specimens with: a) 2n=38, FN=44 from Reserva de Ñacuñán (CMI 7238); b) with 2n=35, FN=45 from Finca La Betania (CMI 7494), extra short arm and secondary constrictions are in boxes; and c) 2n=33, FN=44 from 30 km S
Despite this general pattern, there are several differences among populations. In the Reserva de Ñacuñán only individuals with 2n = 38 (N = 5), and a few with 2n = 37 (N = 2) were detected, indicating a low frequency presence of only 1 Rb fusion. Additionally, in 1 individual we observed the presence of an extra short arm in pair 11 (Table 1). In Reserva de Telteca the only individual studied has 2n = 35 and FN = 44 (Table 1). In Cuenca del Maure 2 individuals have 2n = 36 and one has 2n = 35 (Table 1). In Finca La Betania, individuals with 2n = 38, 37 and 35 were detected (Table 1). In the one with 2n = 35, secondary constrictions were observed in pairs 4 and 5, and extra short arm in pair 4 (Fig. 2b). In that with 2n = 37, secondary constrictions were detected in pairs 5 and 6. The individuals from Salar de Pipanaco showed diploid numbers that varied from 2n=38 to 2n=35, and a fundamental number (FN) of 44 in almost all karyotyped animals (Table 1).
The greatest chromosome diversity was detected in the population of 30 km S of Mendoza Capital. Diploid numbers ranged from 2n = 33 to 2n = 37 (Table 1). In this population, 3 Rb translocations were identified. A new fusion is reported, which involves 2 acrocentrics of different size that form a large submetacentric chromosome (Fig. 3c). This rearrangement was found in homozygosis and in heterozygosis. The individual with 2n = 33 presents the new fusion in heterozygosis (Fig. 3c) and the one with 2n = 34 in homozygosis. The conventional karyotypes of the individuals with 2n = 35, 36 and 37 were similar to those described for the other populations. The cytotypes found in the locality of 18 Km NE of Las Catitas were 2n = 36 and FN = 44 (Table 1), and G-banding and meiotic analyses determined that this individual was heterozygous for 2 Rb translocations (Figs. 3a, 3b). Secondary constrictions were observed in pair 6.
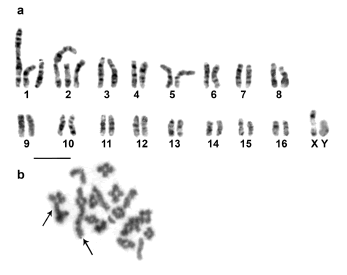
Fig. 3. G-band metaphase (a) and diakinesis cells (b) from an individual of Graomys griseoflavus double heterozygote for Robertsonian translocation from 18 km NE of Las Catitas (CMI 7232). Trivalents are indicated by arrows; XY = sex pair. Scale = 10 mm.
Chromosome measurements
For identifying the pairs involved in the Rb fusions, we compared the chromosome measurements of different cytotypes from different populations (Table 2). The chromosome size of the major fusions described here (pairs Rb 1 and 2) indicated that these rearrangements involved chromosomes 1/6 and 2/5, and appeared to be the same as those found by Zambelli et al. (1994). In the new fusion found at 30 km S of Mendoza Capital, 2n = 33 and 34, the pairs most possibly involved are 3 and 13 or 3 and 14 (Table 2).
Meiotic analysis
Typical trivalents were observed in the meiosis of heterozygote individuals (Fig. 3b). Neither univalents nor other meiotic disturbances were observed. All analyzed trivalents showed the same meiotic behavior in relation to frequency and distri bution of chiasmata, despite having originated from different chromosomes (Fig. 3b). From a total of 20 analyzed trivalents, all had only one chiasma per chromosome arm and 70% of them were distal, 35% interstitial and 5% proximal.
Morphology
Specimens studied here exhibit some variability in dorsal and ventral coloration; dorsally, some specimens are very dark with predominance of black hairs, while others are lighter, with predominance of light brown and yellow hairs. On the lateral side of the body, some specimens have a conspicuous yellowish lateral line that is absent in others. The tail is long and hairy throughout its length and with its tip ending in a tuft of dark hairs in all specimens. Ventral body hairs vary in coloration at the base from completely white (Nw) to gray (Ng) or black (Nb), but being always white at the tip. This character was variable among the 20 specimens from Reserva de Ñacuñán (Nw = 9, Ng = 7, Nb = 4), the 6 from 30 km S of Mendoza Capital (Ng = 4, Nb = 2) and the 6 from Cuenca del Maure (Nw = 3, Ng = 2, Nb = 1). Against this, in Salar de Pipanaco (Nw = 7), Finca La Betania (Nb = 5), Reserva de Telteca, (Nw = 2), Punta de Agua (Nb = 1) and 18 Km NE of Las Catitas (Ng = 1) only one color was observed.
Regarding the cranial measurements, significant differences between males and females of the entire sample (N = 48) were detected in 7 variables by Mann-Whitney U-test. Condylobasal length (p = 0.025), zygomatic breadth (p = 0.030), greatest length of skull (p = 0.011), basal length (p = 0.026), breadth of braincase (p = 0.016), length of bulla with tube (p = 0.004) and palatal bridge (p = 0.026) were significantly higher in males than in females. The same tendency was detected in the other metrical variables. On the other hand, length of the maxillary toothrow and length of mandible toothrow are higher in females than in males (Table 3, supplementary material). A Kruskal Wallis test shows not significant differences among samples from different localities in external and cranial measurements (Table 3, supplementary material). Out of 28 variables, only 1 (weight) shows significant differences among populations.
DISCUSSION
Graomys griseoflavus presents a Robertsonian chromosome polymorphism of wide distribution. The diploid numbers found vary from 2n = 33 to 2n = 38 (Tiranti, 1998; Theiler et al., 1999a; García and Walker, 2004; Rodríguez and Theiler, 2007; this work). Most of the 2n found in this report coincide with that described by other authors, with the exception of the 2n = 33 cytotype from Luján de Cuyo which is described here for the first time. Chromosome complements with 2n = 33, 34, 35, 36 and 37 are related to the cytotype with 2n = 38 through 1, 2 or 3 Rb translocations. This is the first report of a third fusion in G. griseoflavus, and was found in combination with 2 other fusions.
Early studies by Zambelli et al. (1994) through G-banding concluded that the pairs involved in the 2 previously known fusions of G. griseoflavus were pair 1 with 6, and 2 with 5, in individuals from a relatively wide geographic range (Buenos Aires, Catamarca, La Rioja, and Mendoza provinces). In the new fusion presented in this study, the pairs possibly involved are 3 with 13 or 14. The other fusions found appear to correspond to the chromosomes 1/6 and 2/5 presented by Zambelli et al. (1994), although this cannot be fully confirmed due to differences in the G-banded resolution of chromosomes. The karyotypic data from San Luis Province reported fusions between pairs 2 with 12, and 4 with 5; and this involves monobrachial homology with other populations (García and Walker, 2004). The fusion described in this work was found in homo and heterozygote state, which indicated it is not a spontaneous mutant. It is part of the chromosome complement of the population. In some localities (Salar de Pipanaco, 30 km S of Mendoza Capital and Finca La Betania), the observed diploid numbers indicated that there are present more than one fusion in heterozygous state; because otherwise it would have been impossible to find more than one odd diploid number together. However, in previous works no more than one fusion in heterozygosis was found in several populations (Zambelli et al., 1994, 2003; Tiranti, 1998). Additional studies are needed to understand this Rb system of translocations, which is distributed across the whole species' range, and appeared to be more complex than previously suspected.
Graomys griseoflavus is another example where Rb translocations generate chromosomal polymorphism and polytypism. This distribution pattern of Rb variability was also observed in the phyllotine Eligmodontia puerulus, the oryzomyine Holochilus brasiliensis and the murine Mus musculus domesticus (Nachman, 1992; Piálek et al., 2005; Lanzone et al., 2011). The low frequency of the new fusion found in G. griseoflavus and its geographic restriction suggest that it is a recent chromosomal mutation, independently acquired in that population. On the other hand, cytotypes found in Reserva de Ñacuñán were 2n = 38 and 2n =37, which contrasts with the cytotypes found in near sites (Finca La Betania with 2n = 35, 37, 38; 30 km S of Mendoza Capital with 2n = 33-38 and 18 Km NE of Las Catitas with 2n = 36 double heterozygote), suggesting population isolation.
With respect to the origin of the Rb fusions in G. griseoflavus, 2 hypotheses were proposed. One of them postulates a simultaneous and independent occurrence of these mutations in several individuals, without a substantial reduction of population size (Theiler et al., 1999a). The absence of significant bottlenecks in the speciation of G. griseoflavus is supported by its high levels of isozyme heterozygosity; as well as its high levels of nucleotide diversity and similar haplotype diversity to G. chacoensis, in the control region of the mitochondrial genome (Theiler and Gardenal, 1994; Theiler et al., 1999a; Martínez et al., 2010b).
The other hypothesis proposes a single origin for Rb animals, without ruling out the occurrence of a founder event during the chromosomal differentiation (Catanesi et al., 2002; Zambelli et al., 2003). This is supported by the observation that all Rb karyomorphs studied by Catanesi et al. (2002) and by Ferro and Martínez (2009) were grouped in a single clade, while the ancestral G. chacoensis formed a different one. Moreover, a non-random sequence of chromosomal evolution, where occurrence of 1 Rb fusion needs to be preceded by fixation of a different one was proposed by Zambelli et al. (2003). Our data contradict this last hypothesis, because the presence of a double heterozygote from 18 Km NE of Las Catitas was confirmed by G-banding and meiotic analysis. Furthermore, in 4 populations individuals showed more than one fusion in heterozygous state, as can be inferred from diploid numbers. These results indicated that the generation and/or maintenance of a new fusion is not necessarily preceded by fixation of the others. In general, our results support the hypothesis that chromosomal evolution in G. griseoflavus has occurred by multiple independent centric fusions.
Within the genus, the species with the closest karyotype to G. griseoflavus is G. chacoensis, which lacks the Rb fusion found in heterozygosis, as well as 2 fixed fusions found in G. griseoflavus (15/17 and 16/18) (Zambelli et al., 1994). However, a 2n = 41 individual of G. chacoensis was early found, which was interpreted as an independent Rb translocation (Zambelli et al., 1994). These authors, through G-banding, indicated that the chromosome fusion in the 2n = 41 karyotype is 1/6, the same as one found in G. griseoflavus. This suggests that both species are prone to generating this type of mutation in some chromosome pairs, as was suggested for other rodent species (Lanzone et al., 2011). However, it is not clear how many individuals were found with this chromosome constitution. Zambelli et al. (1994) report only one in Deán Funes (Córdoba), which is in concordance with a spontaneous mutant hypothesis. Later, Catanesi et al. (2002), in a study of molecular genetics, reported another individual with 2n = 41 from Chamical (La Rioja), and indicated that the fusion involved is the same (1/6), which suggested that this rearrangement can cover a broader distribution range. But, as G. chacoensis is a poorly studied species and confusion has arisen around the localities of origin of some animals (Ferro and Martínez, 2009), we cannot be sure about whether both species share the same polymorphic rearrangement or whether in G. chacoensis this was de novo generated as a spontaneous mutation.
While Rb translocations are the most common type of chromosomal rearrangement in G. griseoflavus, Zambelli et al. (1994) described the presence of 2 major pericentric inversions in this species. In the specimens studied here, we found no such inversions. But the presence of extra short arms in some individuals, not previously described, indicates occurrence of additional chromosomal variability in this species. The presence of polymorphic inversions was detected in G. chacoensis too (Zambelli et al., 1994; Tiranti, 1998; Lanzone et al., 2007). However, these rearrangements in both species appear to be restricted to few localities. The limited geographic range of these chromosome inversions and its restriction to the region suggested as the original area of phylogenetic divergence between the 2 species (Theiler et al., 1999a; Martínez et al., 2010b) can suggest a genetic instability zone where the speciation process took place. Phylogeographic approaches reinforce the hypothesis that the area of cladogenetic event was the central-western region of the country (La Rioja and Catamarca provinces) (Martínez et al., 2010b). The speciation process was suggested to occur in a parapatric context (Theiler et al., 1999a). However, in this area there are populations of G. chacoensis and G. griseoflavus which are separated by a geographic barrier (Sierras de Ambato) such as populations of Chumbicha and Salar de Pipanaco, indicating that an allopatric context for the cladogentic event cannot be ruled out.
Regarding the role of chromosomes in the speciation process, several models have been postulated (Sites and Moritz, 1987; King, 1993). Theiler et al. (1999a) discuss different models of chromosomal speciation that could fit the observed pattern of chromosomal differentiation between G. griseoflavus and G. chacoensis. But given that Rb heterozygotes apparently have no evident meiotic problems (this study) and because observations of laboratory crosses reveal a complex pattern of pre and post-matting reproductive isolation (Theiler and Blanco, 1996b; Theiler et al., 1999b), the primary role of these rearrangements in the separation of both species is questionable. Maybe a more complex mode of speciation accounts for the split of these taxa, where the genetic differences are accumulated in the chromosomes with structural rearrangement that differentiated both species, as evidenced in the human and chimpanzees lineages (Navarro and Barton, 2003).
At morphological level, G. griseoflavus seems to have some degree of sexual dimorphism. There are 7 metric variables where males are larger than females, and this trend is observed in most measurements. In mammals in general, and in phyllotines in particular, this is the most common type of sexual dimorphism; although the reverse pattern was also seen (Rodríguez et al., 2012 and literature cited there). Aside from sexual differences, G. griseoflavus presents low morphological variability in quantitative characters. Although the samples from different localities are small, morphometric differentiation among population is poor, and this is coincident with the results reported in other studies for this species (Rosi, 1983; Piciucci de Fonollat et al., 1985; this work). In qualitative characters, ventral coloration seems to have three stages clearly defined and differentiation among populations. This ventral character has the same type of variation in the related phyllotine E. puerulus (Lanzone, 2009), but its biological significance as its genetic base is not understood.
At intrageneric level, G. griseoflavus is the species with the highest chromosome variability. Coincidently, in the control region of mitochondrial DNA, in G. griseoflavus were observed more variable sites and higher nucleotide diversity and divergence than in G. chacoensis; although in both species the intra- and inter-specific divergence in this genetic marker is scarce (Martínez et al., 2010b). However in morphology, when the coefficients of variation are calculated from the craniometrical data presented by Martínez et al. (2010a) and compared with that obtained in this study, G. chacoensis appears to be more variable than G. griseoflavus. In conclusion, there is uncoupling among morphological, chromosomal and molecular variability in G. griseoflavus, which appeared to be a frequent pattern in some rodents (Modi, 2003; Lanzone et al., 2011).
ACKNOWLEDGMENTS
The authors wish to thank María Ana Dacar and Silvia Brengio for their cooperation in the laboratory. Our thanks to Nelly Horak for her assistance with the English version. We are grateful to the associated editor Pablo Teta and one anonymous referee for critical review of an early version of this paper. This study was partially funded by Agencia SECYT PICT 25778, 11768 and PIP Consejo Nacional de Investigaciones Científicas y Técnicas CONICET 5944 grants to RAO; PICT 2010/1095 and CONICET PIP 198 to CL.
LITERATURE CITED
1. CATANESI CI, L VIDAL-RIOJA, and A ZAMBELLI. 2006. Molecular and phylogenetic analysis of mitochondrial control region in Robertsonian karyomorphs of Graomys griseoflavus (Rodentia, Sigmodontinae). Mastozoología Neotropical 13:21-30. [ Links ]
2. CATANESI CI, L VIDAL-RIOJA, JV CRISCI, and A ZAMBELLI. 2002. Phylogenetic relationships among Robertsonian karyomorphs of Graomys griseoflavus (Rodentia, Muridae) by mitochondrial cytochrome b DNA secuencing. Hereditas 136:130-136. [ Links ]
3. D'ELÍA G. 2003. Phylogenetics of Sigmodontinae (Rodentia, Muroidea Cricetidae), with special reference to the akodont group, and with additional comments on historical biogeography. Cladistics 19:307-323. [ Links ]
4. EVANS EP, G BRECKON, and CE FORD. 1964. An air-drying method for meiotic preparations from mammalian testes. Cytogenetics 3:289-294. [ Links ]
5. FERRO LI and JJ MARTÍNEZ. 2009. Molecular and morphometric evidences validate a Chacoan species of the grey leaf-eared mice genus Graomys (Rodentia: Cricetidae: Sigmodontinae). Mammalia 73:265-271. [ Links ]
6. FORD CE and JL HAMERTON. 1956. A colchicine, hypotonic citrate, squash sequence for mammalian chromosomes. Stain Technology 31:247-251 [ Links ]
7. GARCÍA AA and L WALKER. 2004. Fusiones céntricas independientes en poblaciones de Graomys griseoflavus (Rodentia: Sigmodontinae). Jornadas Argentinas de Mastozoología 9:85. [ Links ]
8. GÓMEZ-LAVERDE M, RP ANDERSON, and LF GARCIA. 2004. Integrated systematic evaluation of the Amazonian genus Scolomys (Rodentia, Sigmodontinae). Mammalian Biology 69:119-139. [ Links ]
9. HERSHKOVITZ P. 1962. Evolution of Neotropical Cricetine Rodents (Muridae) with special reference to the Phyllotine group. Fieldiana Zoology 46:1-524. [ Links ]
10. KING M. 1993. Species evolution. The role of chromosome change. Cambridge. University Press, Cambridge. [ Links ]
11. LANZONE C. 2009. Sistemática y evolución del género Eligmodontia (Rodentia, Muridae, Sigmodontinae). Tesis de Doctorado. Universidad de Córdoba, Facultad de Ciencias Exactas, Físicas y Naturales. Ciudad de Córdoba, Argentina. [ Links ]
12. LANZONE C, RA OJEDA, and MH GALLARDO. 2007. Integrative taxonomy, systematics and distribution of the genus Eligmodontia (Rodentia, Cricetidae, Sigmodontinae) in the temperate Monte Desert of Argentina. Zeitschrift für Säugetierkunde 72:299-312. [ Links ]
13. LANZONE C, A OJEDA, RA OJEDA, S ALBANESE, D RODRIGUEZ, and MA DACAR. 2011. Integrated analyses of chromosome, molecular and morphological variability in the sister species of Andean mice Eligmodontia puerulus and E. moreni (Rodentia, Cricetidae, Sigmodontinae). Mammalian Biology 76:555-562. [ Links ]
14. MARTIN RE, RH PINE, and AF DEBLASE. 2001. A manual of mammalogy with keys to families of the world. McGraw-Hill Companies, New York. [ Links ]
15. MARTÍNEZ JJ and V DI COLA. 2011. Geographic distribution and phenetic skull variation in two close species of Graomys (Rodentia, Cricetidae, Sigmodontinae). Zoologischer Anzeiger 250:175-194. [ Links ]
16. MARTÍNEZ JJ, JM KRAPOVICKAS, and GR THEILER. 2010a. Morphometrics of Graomys (Rodentia, Cricetidae) from Central-Western Argentina. Mammalian Biology 75:180-185. [ Links ]
17. MARTÍNEZ JJ, RE GONZÁLEZ-ITTIG, GR THEILER, RA OJEDA, C LANZONE, A OJEDA, and CN GARDENAL. 2010b. Patterns of speciation in two sibling species of Graomys (Rodentia, Cricetidae) based on mtDNA sequences. Journal of Zoological Systematics and Evolutionary Research 48:159-166. [ Links ]
18. MODI WS. 2003. Morphological, chromosomal, and molecular evolution are uncoupled in pocket mice. Cytogenetics and Genome Research 103:150-154. [ Links ]
19. MUSSER GG and D CARLETON 2005. Subfamily Sigmodontinae. Pp. 1086-1186, in: Mammal Species of the World, A Taxonomic & Geographic Reference (DE Wilson and DA Reeder, eds.). 3rd Edition, The John Hopkins University Press, Maryland. [ Links ]
20. NACHMAN MW. 1992. Meiotic studies of Robertsonian polymorphisms in the South American marsh rat, Holochilus brasiliensis. Cytogenetics and Cell Genetics 61:17-24. [ Links ]
21. NAVARRO A and NH BARTON. 2003. Chromosomal speciation and molecular divergence - Accelerated evolution in rearranged chromosomes. Science 300:321-324. [ Links ]
22. PATTON JL. 1967. Chromosome studies of certain pocket mice, genus Perognathus (Rodentia, Heteromyidae). Journal of Mammalogy 48:27-37. [ Links ]
23. PEARSON OP and JL PATTON. 1976. Relationship among South American phyllotine rodents based on chromosomal analysis. Journal of Mammalogy 57:339-350. [ Links ]
24. PIÁLEK J, HC HAUFFE, and JB SEARLE. 2005. Chromosomal variation in the house mouse: Review. Biological Journal of the Linnean Society 84:535-563. [ Links ]
25. PICIUCCI DE FONOLLAT AM, G SCROCCHI, MR AGUIRRE, and JL ORGEIRA. 1985. Aportes a la morfometría de Graomys griseoflavus (Rodentia: Cricetidae). Historia Natural 5:225-225 [ Links ]
26. REIG OA. 1986. Diversity patterns and differentiation of high Andean rodents. Pp. 404-439, in: High Altitude Tropical Biogeography (F Vuilleumier and M Monasterio, eds.). Oxford University Press, New York. [ Links ]
27. RODRÍGUEZ V and GH THEILER. 2007. Micromamíferos de la región de Comodoro Rivadavia (Chubut, Argentina). Mastozoología Neotropical 14:97-100. [ Links ]
28. RODRÍGUEZ D, C LANZONE, V CHILLO, PA CUELLO, S ALBANESE, AA OJEDA, and RA OJEDA. 2012. Historia natural de un roedor raro del desierto argentino, Salinomys delicatus (Cricetidae: Sigmodontinae). Revista Chilena de Historia Natural 85:13-27. [ Links ]
29. ROSI MI. 1983. Notas sobre la ecología, distribución y sistemática de Graomys griseoflavus griseoflavus (Waterhouse, 1837) (Rodentia, Cricetidae) en la provincia de Mendoza. Historia Natural 3:161-167. [ Links ]
30. SEABRIGHT M. 1971. A rapid banding technique for human chromosomes. Lancet 2:971-972. [ Links ]
31. SIKES RS, EC MONJEAU, CJ PHILLIPS, and JR HILLYARD. 1997. Morphological versus chromosomal and molecular divergence in two species of Eligmodontia. Zeitschrift für Säugetierkunde 62:265-280. [ Links ]
32. SITES JW and C MORITZ. 1987. Chromosomal evolution and speciation revisited. Systematic Biology 36:153-174. [ Links ]
33. SMITH MF and JL PATTON. 1993. The diversification of South American murid rodents: Evidence from mitochondrial DNA sequence data for the akodontine tribe. Biological Journal of the Linnean Society 50:149-177. [ Links ]
34. SMITH MF and JL PATTON. 1999. Phylogenetic relationships and the radiation of sigmodontine rodents in South America: Evidence from cytochrome b. Journal of Mammalian Evolution 6:89-128. [ Links ]
35. SPOTORNO AE, LI WALKER, SV FLORES, M YEVENES, JC MARÍN, and C ZULETA. 2001. Evolución de los filotinos (Rodentia, Muridae) en los Andes del Sur. Revista Chilena de Historia Natural 74:151-166. [ Links ]
36. STEPPAN SJ, RM ADKINS, and J ANDERSON. 2004. Phylogeny and Divergence-Date estimates of rapid radiation in muroid rodents based on multiple genes. Systematic Biology 53:533-553 [ Links ]
37. THEILER GR and A BLANCO. 1996a. Patterns of evolution in Graomys griseoflavus (Rodentia: Muridae). II. Reproductive isolation between cytotypes. Journal of Mammalogy 77:776-784. [ Links ]
38. THEILER GR and A BLANCO. 1996b. Patterns of evolution in Graomys griseoflavus (Rodentia, Muridae). III. Olfactory discrimination as a premating isolation mechanism between cytotypes. Journal of Experimental Zoology 274:346-350. [ Links ]
39. THEILER GR and CN GARDENAL. 1994. Patterns of evolution in Graomys griseoflavus (Rodentia, Cricetidae). I. Protein polymorphism in populations with different chromosome number. Hereditas 120:225-229. [ Links ]
40. THEILER GR, CN GARDENAL, and A BLANCO. 1999a. Patterns of evolution in Graomys griseoflavus (Rodentia, Muridae) IV. A case of rapid speciation. Journal of Evolutionary Biology 12:970-979. [ Links ]
41. THEILER GR, RH PONCE, RE FRETES, and A BLANCO. 1999b. Reproductive barriers between the 2n=42 and 2n=36-38 cytotype of Graomys (Rodentia, Muridae). Mastozoología Neotropical 6:129-133. [ Links ]
42. TIRANTI SI. 1998. Cytogenetics of Graomys griseoflavus (Rodentia, Sigmodontinae) in Central Argentina. Zeitschrift für Säugetierkunde 63:32-36. [ Links ]
43. ZAMBELLI A and L VIDAL-RIOJA. 1995. Molecular analysis of chromosomal polymorphism in the South American cricetid, Graomys griseoflavus. Chromosome Research 3:361-367. [ Links ]
44. ZAMBELLI A and L VIDAL-RIOJA. 1996. Loss of nucleolar organizer regions during chromosomal evolution in the South American cricetid Graomys griseoflavus. Genetica 98:53-57. [ Links ]
45. ZAMBELLI A and L VIDAL-RIOJA. 1999. Molecular events during chromosomal divergence of the South American rodent Graomys griseoflavus. Acta Theriologica 44:345-352. [ Links ]
46. ZAMBELLI A, CI CATANESI, and L VIDAL-RIOJA. 2003. Autosomal rearrangements in Graomys griseoflavus (Rodentia): a model of non-random Robertsonian divergence. Hereditas 139:167-173. [ Links ]
47. ZAMBELLI A, L VIDAL-RIOJA, and R WAINBERG. 1994. Cytogenetic analysis of autosomal polymorphism in Graomys griseoflavus (Rodentia, Cricetidae). Zeitschrift für Säugetierkunde 59:4-20. [ Links ]
Studied specimens. Catalog numbers correspond to the Colección Mastozoológica del IADIZA (Instituto Argentino de Investigaciones de Zonas Áridas): CMI; 1 = specimens used only for morphology, 2 = specimens used only for chromosome analysis.
Provincia de Catamarca: Departamento Andalgalá, Salar de Pipanaco (6822, 6908, 07157, 07158, 7184, 7202, 7208).
Provincia de Mendoza: Departamento Santa Rosa, Reserva de Ñacuñán (20171, 22161, 25151, 25621, 25701, 25731, 25741, 61141, 62791, 62821, 62851, 68242, 68911, 68941, 7181, 7190, 71911, 7237, 7238, 72392, 7282, 73021), 18 km NE de Las Catitas sobre Ruta 153 (7232); Departamento Luján de Cuyo, 30 km S de Mendoza Capital (7277, 7278, 7279, 7280, 7281, 7283); Departamento General Alvear, Cochicó, Finca La Betania (7143, 71592, 71821, 71831, 7185, 7247, 74942); Departamento Godoy Cruz, Cuenca del Maure (61071, 61111, 61121, 7286, 7288, 7290); Departamento Lavalle, Reserva de Telteca (47031, 6825); Departamento San Rafael, Punta de Agua (73541)
ONLINE SUPPLEMENTARY MATERIAL
Table 3. Descriptive statistics (mean ± SD, range and coefficient of variation) for external and cranial variables for specimens of Graomys griseoflavus.
http://www.sarem.org.ar/wp-content/uploads/2014/06/SAREM_MastNeotrop_21-1_Lanzone-sup1.doc














