Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
Mastozoología neotropical
Print version ISSN 0327-9383On-line version ISSN 1666-0536
Mastozool. neotrop. vol.25 no.1 Mendoza June 2018
ARTÍCULO
A cost-effective method for rapid identification of the southern muriqui (Brachyteles arachnoides): a contribution for the control of illegal bushmeat trade
M. Luiza Valões Cardoso1, Paula B. Ferreira1, Artur M. Wanderley1, Rodrigo A. Torres2, Mauricio Talebi Gomes3, Rodrigo H. F. Teixeira4, 5, José M. B. Duarte6 and José E. Garcia1
1 Centro Acadêmico de Vitória, Universidade Federal de Pernambuco, Vitória de Santo Antão, Brazil [Correspondence: José E. Garcia <jegarcia30@gmail.com>]
2 Departamento de Zoologia, Universidade Federal de Pernambuco, Recife, Brazil
3 Departamento de Ciências Ambientais, Universidade Federal de São Paulo, Campus Diadema, Diadema, São Paulo, Brazil
4 Parque Zoológico Municipal Quinzinho de Barros, Sorocaba, São Paulo, Brazil
5 Programa de Animais Selvagens, Universidade Estadual Paulista "Júlio de Mesquita Filho", Botucatu, São Paulo, Brazil
6 Departamento de Zootecnia, Universidade Estadual Paulista "Júlio de Mesquita Filho", Jaboticabal, São Paulo, Brazil
Recibido 27 junio 2017.
Aceptado 8 dic 2017.
Editor asociado: R González-Ittig
ABSTRACT.
To control illegal wildlife-product trade and protect endangered species of animals, unambiguous identification of the captured specimens is essential. Forensic genetic tools have contributed to identify animal species for conservation purposes promoting accurate results for informing public policies and management of the biodiversity. The southern muriqui (Brachyteles arachnoides) is the largest non-human primate of the Neotropical region and is critically endangered (IUCN Red List of Threatened Species), mainly due to the illegal hunting for bushmeat. In this study, we describe a molecular method using PCR/RFLP to differentiate between bushmeat of southern muriqui and the meat of the domestic animals most commonly consumed in Brazil (Bos taurus, Ovis aries, Capra hircus, and Sus scrofa). The method is based on the amplification and digestion with BanI restriction enzyme of the 16S mtDNA region. We also examine 16S mtDNA sequences of the southern muriqui and other 13 sympatric and parapatric wild species of mammals also hunted for bushmeat to examine whether homologies of the BanI restriction sites could lead to species misidentification. The results indicate the utility of this tool as it represents a simple and cost-effective method to differentiate southern muriqui samples from those of the examined domestic and wild sympatric and parapatric species. We hope this molecular tool will help public authorities in crime prevention, and enhance law reinforcement of illegal hunting of threatened animal species.
RESUMO.
Um método rápido e simples para a identificação do muriqui-do-sul (Brachyteles arachnoides): uma contribuição para o combate à caça ilegal.
Para controlar o comércio ilegal de vida selvagem e proteger espécies animais ameaçadas, é essencial a identificação inequívoca dos espécimes capturados. Ferramentas de genética forense têm contribuído na identificação de animais para fins conservacionistas, fornecendo resultados precisos utilizados em políticas de gestão da biodiversidade. O muriqui-do-sul (Brachyteles arachnoides) é o maior primata não humano da região neotropical e está criticamente em perigo (Lista Vermelha da IUCN) devido, principalmente, à caça ilegal para consumo de sua carne. Nesse estudo, descrevemos um método molecular baseado em PCR/RFLP para diferenciar a carne do muriqui-do-sul da carne dos animais domésticos mais consumidos no Brasil (Bos taurus, Ovis aries, Capra hircus, and Sus scrofa). Este método é baseado na amplificação e digestão da região 16S mtDNA com a enzima de restrição BanI. Além disso, sequências da região do 16S mtDNA do muriqui-do-sul e de 13 espécies de mamíferos simpátricos e parapátricos também caçados por sua carne foram examinadas para avaliar a presença de sítios de restrição homólogos que pudessem comprometer a identificação do muriqui-do-sul. Os resultados indicam a utilidade desta ferramenta por representar um método simples e de baixo custo que permite diferenciar amostras de muriqui-do-sul das amostras das espécies domésticas e selvagens que foram examinadas. Esperamos que esta ferramenta molecular ajude as autoridades na prevenção de crimes contra a biodiversidade e cumprimento da legislação contra a caça ilegal de animais ameaçados.
Key words: Brachyteles arachnoides; Forensic genetics; Illegal hunting; Threatened species; 16S mtDNA.
Palavras chave: Brachyteles arachnoides; Caça ilegal; Espécies ameaçadas; Genética forense; 16S mtDNA.
INTRODUCTION
To control illegal wildlife-product trade and protect endangered species of animals, unambiguous identification of seized bushmeat is essential. Specific legislation and treaties controlling the bushmeat trade, such as the Convention on the International Trade of Endangered Species (CITES), depends on taxonomic identification at the species level of suspected illegal products. Forensic genetic approaches have contributed to identify threatened animal species and to inform public policies and management of the observable effects of population declines in many plant and animal species (Jerusalinsky et al. 2011). Animal products are often processed, hampering accurate taxonomic identification based on morphological characters. In such cases, more accurate tools are required to better control the bushmeat trade (Malisa et al. 2006; Eaton et al. 2010).
Databases comprising species-specific DNA sequences of particular gene fragments are valuable for an accurate taxonomic identification of bushmeat products when morphological diagnostic features such as skins, body parts or whole animals are not available (Ross et al., 2003; Malisa et al., 2006; Ratnasingham & Hebert 2007; Eaton et al. 2010). The most common DNA sequences used for the identification of unknown biological samples are mitochondrial genes and regions, such as cytochrome b, cytochrome c oxidase I , the D-loop region, 12S rRNA, and 16S mtDNA (Prakash et al. 2000; Hsieh et al. 2001; Dalebout et al. 2004; Balitzki-Korte et al. 2005; Rönn et al. 2009).
The 16S mtDNA region contains nucleotide variation suitable for its use in essays of digestion of PCR products with restriction enzymes (PCR/RFLP). The selection of specific restriction enzymes for PCR/RFLP analysis allows cost-effective identification of species from DNA samples, making it possible to differentiate morphologically indistinguishable illegal bushmeat from legal products (Bravi et al. 2004).
Study subjects
With approximately 10-12 kg of body weight, the southern muriqui, Brachyteles arachnoides (Geoffroy, 1806), is the largest non-human primate of the New World. It is an herbivorous arboreal monkey, listed as critically endangered by the IUCN Red List of Threatened Species, with less than 1300 individuals remaining within its natural habitat (Mendes et al. 2008; Talebi 2013), the Atlantic forest, a biodiversity hotspot for conservation priorities (Myers et al. 2000). Beyond the ubiquitous fragmentation process under way in the Atlantic forest (Myers et al. 2000), hunting by local human populations is still a major threat to the southern muriqui (Mendes et al. 2008; Talebi 2013).
Although part of the remaining southern muriqui populations are located in protected areas such as the Carlos Botelho State Park (SP) (Fig. 1), hunting pressures to this species have not been substantially reduced. It has been estimated that the current population of the southern muriqui could undergo a 20% decline and a probability of extinction of 50% within the next 50 years, or in the next few generations (Mendes et al. 2008; Talebi 2013).
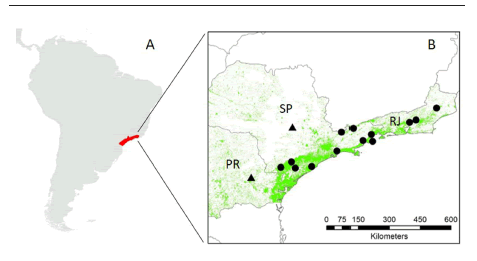
Fig. 1. (A) Historic geographic distribution of the southern muriqui (in red). (B) current sub-populations of the southern muriqui in protected (circles) and non-protected (triangles) areas over the remaining Atlantic forest (in green) in the Brazilian states of Rio de Janeiro (RJ), São Paulo (SP) and Paraná (PR).
The goal of this study was to develop a reliable and cost-effective molecular tool, based on restriction enzyme digestion of the 16S mtDNA region, to differentiate illegal processed bushmeat of the southern muriqui from meat samples of four domestic species broadly consumed in Brazil and 13 species of wild mammals that are sympatric and parapatric with the southern muriqui, which are also hunted for bushmeat. We expect this molecular tool will increase the available information for public authorities and crime prevention to enhance law enforcement against illegal hunting of the endangered southern muriqui.
METHODS
Biological samples
Blood samples of six individuals of the southern muriqui were provided by the Zoo of Sorocaba county (Parque Zoológico Municipal Quinzinho de Barros), in São Paulo State, Brazil, during the zoo's routine medical examination of the captive animals. In addition, muscle samples (one sample per species) from Bos Taurus (cattle), Ovis aries (sheep), Capra hircus (goat), and Sus scrofa (pig), all domestic animals commonly consumed for meat in Brazil, were obtained from public markets in this country.
Laboratorial techniques
DNA isolation from all samples of the southern muriqui and the four domestic species was performed using the DNA Qiagen DNeasy Blood & Tissue kit (Qiagen Inc.), following manufacturer's instructions. DNA quantification was performed using 1% agarose gel electrophoresis, staining DNA samples with GelRedTM (Biotium, Hayward, CA, USA). The mtDNA 16S gene of all the southern muriqui and the four domestic species was amplified by PCR using the universal primers 16Sar-L (5´-CGCCTGTTTTCAAAAACAT-3´) and 16Sbr-H (5´-CGTCTGAACTCAGATCACGT-3´) (Palumbi et al. 1991). The reaction condition contained: 20 ng of DNA, 3.0 mM MgCl2, 1 mM each of dNTP, 1 mM each primer, 0.5 U of Taq DNA polymerase (LGC/ Brazil) and 10X Amplification Buffer (Mg+ free). The reaction started with denaturation at 95 °C for 60 s, followed by 27 cycles of denaturation at 93 °C for 45 s, annealing at 60 °C for 60 s, and extension at 72 °C for 90 s. Finally, there was a hold period of 10 min at 72 °C.
Restriction enzyme digestion
PCR products were digested with 2 U of BanI [restriction site: 5'-G▼GYRCC-3', 3'-CCRYG▲G-5'], according to the manufacturer's instructions (New England BioLabs® Inc.). We selected BanI to digest our PCR products by contrasting our 16S mtDNA sequences (see DNA sequencing below) with 16S mtDNA sequences of other taxa available from GenBank (National Center of Biotechnology Information - NCBI) using Cleaver (Jarman 2006). For a given DNA region (16S mtDNA in this study), Cleaver allows to identify among a range of restriction enzymes, which renders species-specific DNA fragment restriction patterns. For species identification, separation of restriction fragments was accomplished by horizontal 2% agarose gel electrophoresis stained with GelRedTM. To confirm the lack of homology of the BanI restriction sites between the southern muriqui and the four domestic species examined, resulting in different electrophoresis gel band patterns (see Results), we sequenced the 16S mtDNA of our six southern muriqui samples. Sequencing was perfomerd at Macrogen facilities (www.macrogen. com) (Korea) under BigDyeTM terminator cycling conditions on an ABI 3730 xl automatic sequencer (Applied Biosystems, USA). Then we compared these sequences with 16S mtDNA sequences of the four domestic species, which were retrieved from GenBank. The accession numbers of the sequences of the domestic animals obtained in GenBank and used in this analysis were: AF492351.1, for cattle; NC_001941.1, for sheep; NC_005044.1, for goat; and AY334492.2, for pig. For this comparison, we also used one 16S mtDNA sequence of the southern muriqui (DQ078115.1) available in GenBank.
To evaluate whether this method could also distinguish DNA samples of the southern muriqui from other wild mammals with overlapping (sympatric) or adjacent (parapatric) geographic ranges, which are also hunted for bushmeat, we retrieved 16S mtDNA sequences for all species available in GenBank attending to these criteria. Then we compared these sequences to the southern muriqui sequences to look for homologies in the BanI restriction sites. The species available in Genbank used in this analysis, and their accession numbers, are Alouatta caraya (accession number: KC757384.1), A. guariba clamitans (KY202428.1), Callithrix penicillata (KR817256.1), Cebus xanthosternos (KC757410.1), Leontopithecus rosalia (KC757399.1); Mazama americana (JN632657.1), M. gouazoubira (KJ772514.1); Nasua nasua (HM106331.1); Dasypus novemcinctus (KF799981.1); Tamandua tetradactyla (NC_004032.1); Myrmecophaga tridactyla (KT818549.1); Pecari tajacu (AP003427.1); and Hydrochoerus hydrochaeris (AF069533.1). This list of species includes six of the nine genera or species of mammals (deer, South American coati, armadillo, giant anteater, peccary and capybara) for which bushmeat seizures were reported by environmental military police operating between 2013 and 2014 within the distribution range of the southern muriqui (see Chagas et al., 2015).
RESULTS
The sequence of the 16S mtDNA region amplified by PCR was 523 bp in length for the southern muriqui and the four domestic species examined. The BanI enzyme restriction did not digest the 16S mtDNA region of any of the domestic species examined, allowing the identification of the southern muriqui (Fig. 2). In contrast, the electrophoretic pattern after the BanI digestion of the 16S mtDNA region of the southern muriqui resulted in two fragments of 204 bp and 264 bp (Fig. 2).
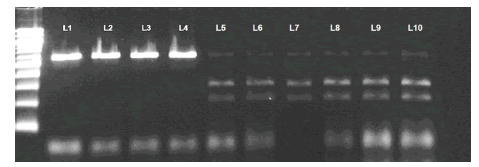
Fig. 2. Gel electrophoresis with 100 bp ladder showing fragments of 16S mtDNA PCR/RFLP for domestic species (L1-L4) and for the southern muriqui (L5-L10). L1-L4 are Bos indicus, Capra hircus, Sus scrofa and Ovis aries, respectively. L5-L10 were digested with BanI enzyme restriction obtaining the following products (204+264 bp).
The 16S mtDNA sequencing of three of our six southern muriqui samples resulted in high quality base calls (Phred quality scores ≥ 20). Our 16S mtDNA sequences of the southern muriqui (GenBank accessions numbers: MH084742, MH084743 and MH084744), as well as the one obtained in GenBank, showed BanI restriction sites [5'-G▼GYRCC-3] between positions 56 and 61 and 265 and 270 (Fig. 3). This reveals that the two fragments of 204 bp and 264 bp observed in the agarose gel for the southern muriqui samples (Fig. 2) are the product of digestion of the BanI restriction site between positions 265 and 270. In addition, a small fragment of 55 pb is also expected as the product of digestion of the restriction site between positions 56 and 61. However, this fragment could not be visualized by agarose gel electrophoresis given its small size. None of the four domestic species shared these restriction sites with the southern muriqui (Fig. 3), which was expected as their PCR products were not digested by BanI. Among the examined sympatric and parapatric wild mammals, the restriction site observed for the southern muriqui in positions 56-61 of the 16S mtDNA region was shared only with the other primate species examined and with Dasypus novemcinctus, whereas the restriction site observed in positions 265-269 was exclusive to the southern muriqui (Fig. 3). Therefore, the DNA fragment restriction pattern expected for the species sharing only the restriction site between nucleotide positions 56 and 61 (one fragment of 55 bp [not visible in agarose gel] and another of 466 bp) is different than the restriction pattern observed for the southern muriqui.
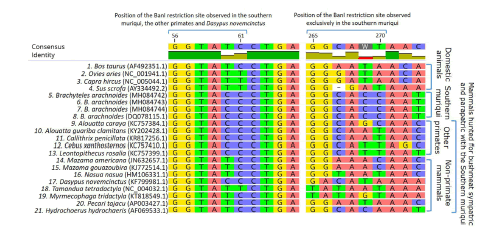
Fig. 3. Multiple alignment of polymorphic sequences of the 16S mtDNA region from four domestic species raised for meat production in Brazil; the southern muriqui and its sympatric and parapatric species of primates, deer, and coati, which are hunted for bushmeat. BanI restriction enzyme site: 5'-G▼GYRCC-3'. GenBank accession numbers of the sequences are within brackets.
In sum, the digestion of the 16S mtDNA region with the BanI restriction enzyme allows us to discriminate between samples of the southern muriqui from those of the four domestic species commonly raised for meat consumption in Brazil, as well as from those of 13 sympatric and parapatric species of mammals examined. All the domestic and wild species used in this study, which sequences from multiple accessions were available in GenBank showed no intraspecific variation in the BanI restriction sites. Thus, misidentification between the southern muriqui and these species using the method here described is unlikely.
DISCUSSION
The findings of this study suggest a great potential for the use of the BanI restriction enzyme as a simple and cost-effective forensic tool to identify southern muriqui bushmeat, even for highly processed samples. Claims by suspected hunters that samples belong to other species can be discredited. There are currently no sequences of the relevant 16S region of the lowland paca (Cuniculus paca), opossum (Didelphis) and tapeti (Sylvilagus brasiliensis), local species that have been seized by environmental military police (Chagas et al., 2015) within the southern muriqui geographic range. The expected restriction pattern for these species could therefore not be examined. Nonetheless, it is very unlikely these distantly related taxa invalidate our method due to shared BanI restriction sites in the 16S mtDNA region with the southern muriqui. The 16S mtDNA is highly conserved within animal species and, thus, has been used for identification of a wide range of mammals, including New World monkeys (Horovitz & Meyer 1995; Sarri et al. 2014).
No 16S mtDNA sequences of the allopatric northern muriqui (Brachyteles hypoxanthus) are available in GenBank, and we were unable to have access to northern muriqui individuals to perform DNA extraction given its rarity. Until data become available for the northern muriqui, we recommend the use of the forensic tool here described exclusively within the boundaries of the southern muriqui geographic range.
CONCLUSIONS
This study demonstrates the existence of nucleotide variation in the 16S mtDNA region that are likely useful for the molecular identification of the southern muriqui by means of PCR/RFLP, a very simple and cost-effective method. The use of this molecular tool is promising in forensic genetics and may help public authorities fighting against bushmeat trade of the southern muriqui.
ACKNOWLEDGEMENTS
The authors are thankful to Quinzinho de Barros Zoo/ Sorocaba's Zoo, in Sorocaba County, São Paulo State, Brazil (Parque Zoológico Municipal Quinzinho de Barros/Zoológico de Sorocaba) for providing the southern muriqui blood samples used in the study; and to CNPq (Conselho Nacional de Desenvolvimento Cientifico e Tecnológico), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) and FACEPE (Fundação de Amparo à Ciência e Tecnologia de Pernambuco) for financial support. RAT and JMBD are grateful to CNPq for researcher fellowships.
LITERATURE CITED
1. Balitzki-Korte, B., K. Anslinger, C. Bartsch, & B. Rolf. 2005. Species identification by means of pyrosequencing the mitochondrial 12S rRNA gene. International Journal of Legal Medicine 119:291-294. [ Links ]
2. Bravi, C. M., J. P. Lirón, P. M. Mirol, M. V. Ripoli, P. Peral-García, & G. Giovambattista. 2004. A simple method for domestic animal identification in Argentina using PCR-RFLP analysis of cytochrome b gene. Legal Medicine 6:246-251. [ Links ]
3. Chagas, A. T. A., M. A. Costa, A. P. V. Martins, L. C. Resende, & E. Kalapothakis. 2015. Illegal hunting and fishing in Brazil: a study based on data provided by environmental military police. Natureza & Conservação (Brazilian Journal of Nature Conservation) 13:183-189. [ Links ]
4. Dalebout, M. L., C. S. Baker, J. G. Mead, V. G. Cockcroft, & T. K. Yamada. 2004. A comprehensive and validated molecular taxonomy of beaked whales, family Ziphiidae. Journal of Heredity 95:459-473. [ Links ]
5. Eaton, M. J., G. L. Meyers, S. O. Kolokotronis, M. S. Leslie, A. P. Martin, & G. Amato. 2010. Barcoding bushmeat: molecular identification of Central African and South American harvested vertebrates. Conservation Genetics 11:1389-1404. [ Links ]
6. Horovitz, I., & A. Meyer. 1995. Systematics of New World monkeys (Platyrrhini, Primates) based on 16S mitochondrial DNA sequences: a comparative analysis of different weighting methods in cladistic analysis. Molecular Phylogenetics and Evolution 4:448-456. [ Links ]
7. Hsieh, H. M. et al. 2001. Cytochrome b gene for species identification of the conservation animals. Forensic Science International 122:7-18. [ Links ]
8. Jarman, S. N. 2006. Cleaver: software for identifying taxon specific restriction endonuclease recognition sites. Bioinformatics 22:2160-2161. [ Links ]
9. Jerusalinsky, L., M. Talebi, & F. R. Melo. 2011. Plano de ação nacional para a conservação dos muriquis. Brasília. [ Links ]
10. Malisa, A., P. Gwakisa, S. Balthazary, S. Wasser, & B. Mutayoba. 2006. The potential of mitochondrial DNA markers and polymerase chain reaction-restriction fragment length polymorphism for domestic and wild species identification. African Journal of Biotechnology 5:1588-1593. [ Links ]
11. Mendes, S. L., M. M. Oliveira, R. A. Mittermeier, & A. B. Rylands. 2008. Brachiteles arachnoides. The IUCN Red List of Threatened Species. [ Links ]
12. Myers, N., R. A. Mittermeier, C. G. Mittermeier, G. A. B. da Fonseca, & J. Kent. 2000. Biodiversity hotspots for conservation priorities. Nature 403:853-858. [ Links ]
13. Palumbi, S., S. Romano, W. O. Mcmillan, & G. Grabowski. 1991. The simple fool's guide to PCR, version 2. University of Hawaii Press Honolulu. [ Links ]
14. Prakash, S., M. S. Patole, S. V. Ghumatkar, S. K. Nandode, B. M. Shinde, & Y. S. Shouche. 2000. Mitochondrial 12S rRNA sequence analysis in wildlife forensics. Current Science 78:1239-1241. [ Links ]
15. Ratnasingham, S., & P. D. N. Hebert. 2007. BOLD: The Barcode of Life Data System: Barcoding. Molecular Ecology Notes 7:355-364. [ Links ]
16. Rönn, A. C. et al. 2009. First generation microarray-system for identification of primate species subject to bushmeat trade. Endangered Species Research 9:133-142. [ Links ]
17. Ross, H. A. et al. 2003. DNA Surveillance: web-based molecular identification of whales, dolphins, and porpoises. Journal of Heredity. [ Links ]
18. Sarri, C. et al. 2014. A new set of 16S rRNA universal primers for identification of animal species. Food Control 43:35-41. [ Links ]
19. Talebi, M. G. 2013. Genus Brachyteles. Handbook of the Mammals of the World, v. 3. Lynx Edicions, Barcelona, 7 pp. [ Links ]














