Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
Biocell
Print version ISSN 0327-9545
Biocell vol.30 no.2 Mendoza May/Aug. 2006
Evaluation of a multiplex PCR method to detect enteroaggregative Escherichia coli
M.E. Rüttler, C.S. Yanzón, M.J. Cuitiño, N.F. Renna, M.A. Pizarro and A.M. Ortiz.
Área de Química Biológica, Facultad de Ciencias Médicas, Universidad Nacional de Cuyo. Mendoza, Argentina.
Address correspondence to: Dra. María Elena Rüttler. Área de Química Biológica, Facultad de Ciencias Médicas, Universidad Nacional de Cuyo. Casilla de Correo 33, (5500) Mendoza, ARGENTINA. E-mail: mruttler@fcm.uncu.edu.ar
Abstract: Enteroaggregative Escherichia coli (EAEC) has been implicated in sporadic diarrhea in children and adults and has been identified as the cause of several outbreaks worldwide. The HEp-2 test remains the gold standard for identification of this pathotype. A 60-65 MDa plasmid encodes the aggregative adherence fimbriae (AAF/I and AAF/II), a transcriptional activator ( aggR gene), the enteroaggregative heat-stable enterotoxin EAST1 ( astA gene) and a cytotoxin (Pet). The standard assay for EAEC is performed only in research laboratories, because it is expensive, labor intensive and time-consuming. The Polymerase Chain Reaction (PCR) offers the possibility of rapid diagnosis. In the current study, a multiplex PCR assay which checks aggR and astA genes was designed. Eigthy- eight E.coli strains, isolated from children with acute diarrhea in Mendoza , Argentina , were characterized by the reference method (HEp-2 assay), and by aggRastA PCR. A strong correlation between the presence of the specific marker aggR and the reference test was found. The astA gene had a similar distribution between aggregative and localized strains, indicating that this gene could not be considered as a marker of EAEC. We conclude that aggR may be used to identify EAEC, using the PCR method as a screening test.
Key words: Diarrhea; Escherichia coli; Cell culture; PCR (Polymerase Chain Reaction).
Introduction
Epidemiological studies done in the past century have demonstrated an association of specific serotypes of Escherichia coli and diarrhea. Strains belonging to these serotypes were referred as Enteropathogenic Escherichia coli (EPEC), but early studies did not identify any pathogenic factor, that could account for their pathogenicity.
Since EPEC adherence to intestinal mucosa in vivo has been demonstrated, initial work attempted to characterize adhesive mechanisms. In addition, the development of in vitro human-epithelial cell adherence tests demonstrated the correlation between some phenotype and the pathogenicity of EPEC. (Cravioto et al. , 1979; Cravioto et al. , 1991; Law, 1994; Nataro et al., 1987).
With the HEp-2 or Hela cell assay three distinct patterns of adherence have been described: Localized Adherence (LA) in which bacteria form characteristic microcolonies on the surface of the HEp-2 cell (Fig. 1), Diffuse Adherence (DA), in which bacteria adhere evenly to the whole cell surface (Fig. 2), and aggregative adherence, in which aggregated bacteria attach to the cell in a stacked-brick arrangement (Fig. 3). Thus, this test allows to differenciate among all three adherent diarrheigenic categories: EPEC, Diffuse Adherence Escherichia coli (DAEC) and Enteroaggregative Escherichia coli (EAEC).
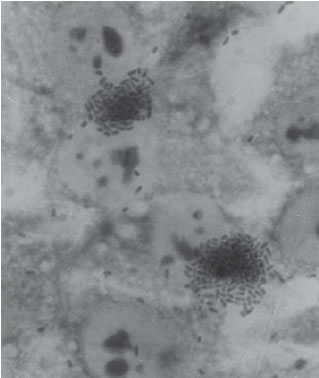
FIGURE 1. Localized Adherence (LA) typical of EPEC. Bacteria form characteristic microcolonies on the surface of the HEp-2 cell. (X 1,000)
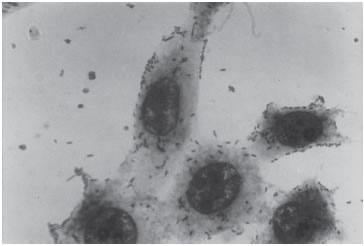
FIGURE 2. Diffuse adherence (DA) which defines DAEC. Bacteria are dispersed over the cell surface. (X 1,000)
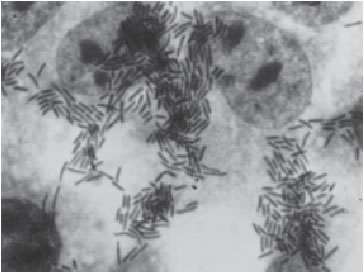
FIGURE 3. Aggregative Adherence (AA), which defines EAEC. Bacteria attach to both cells and coverslips in a stacked brick appearance. (X 1,000)
The LA phenotype is associated to the induction of the attaching and effacing lesions (A/E) produced by EPEC. The most notable feature of the epidemiology of disease due to EPEC is the striking age distribution, seen in people infected with this pathogen. EPEC infection is primarily a disease of infants younger than 2 years, with a strongest correlation in infants younger than 6 months. In children older than 2 years, EPEC can be isolated from healthy and sick individuals, but a statistically significant correlation with disease is usually not found (Nataro and Kaper, 1998). The pathogenic role of E. coli showing a DA pattern (DAEC) in the etiology of diarrheal disease is controversial. EAEC is an increasing important cause of diarrhea. E.coli belonging to this category produces watery diarrhea, which is often persistent and can be inflammatory (Bhatnagar et al. , 1993).
EAEC has been implicated in sporadic diarrhea in children and adults and has been identified as the cause of several outbreaks worldwide (Bhan et al ., 1989; Cobeljic et al. , 1996; Nataro et al ., 1998; Okeke and Nataro, 2001). The HEp-2 test remains the gold standard for identification of this pathotype. In at least some strains, adherence is encoded on a 60-65 MDa plasmid (designated pAA) that also encodes several other putative virulence factors. pAA- encoded factors include the aggregative adherence fimbriae (AAF/I and AAF/II), a transcriptional activator ( aggR gene), the enteroaggregative heat-stable enterotoxin EAST1 ( astA gene) and a 104-k-Da cytotoxin designated as Pet (Czeczulin et al ., 1991; Eslava et al. , 1998; Baudry et al ., 1990; Debroy et al. , 1994).
The standard assay for EAEC, which demonstrates aggregative attachment of organisms to HEp-2 cells, is performed only in research laboratories, because it is expensive, labor intensive and time-consuming. With the advent of Polymerase Chain Reaction (PCR), it has become possible to identify pathogenic factors in bacterial isolates, offering the possibility of rapid diagnosis of E.coli infections (Dutta et al ., 1999; Okeke et al ., 2000; Schmidt et al., 1995; Tsai et al., 2003; Yatsuyanagi et al., 2002; Yatsuyanagi et al., 2003).
In a previous study, we evaluated the value of a PCR method as a screening test for the diagnosis of EAEC strains (Rüttler et al., 2002). The virulence factors investigated in that study were AAF/I and EAST1. The sensitivity obtained for AAF/I was low, suggesting that some of the strains possessed other adherent factor, which in the present work was not investigated.
In the current study, we designed a set of primers to identify aggR gene, which contains the sequence for a transcriptional activator common to both AAF/I and AAF/II, in order to improve the sensitivity previously obtained, when were only checked AAF/I. We included both, aggR and astA gene (EAST1) in a multiplex PCR assay (Elías et al ., 1999; Nataro and Cravioto, 1998; Rich et al., 1999; Nataro et al., 1993).
Materials and Methods
The strains examined in this work were isolated during an epidemiologic study of acute diarrhea in children younger than 2 years old. The present study was performed in the Humberto Notti Children´s Hospital in Mendoza , Argentina , during 1999 and 2000.
Samples were obtained by spontaneous evacuation, collected in sterile recipients and processed within two hours. Direct microscopic exam was performed on the stool with Gram stain, Blue Methylene stain (leukocytes observation) and fresh exam in order to detect Entamoeba histolytica and Giardia lamblia . To identify cryptosporidium, modified Ziehl – Neelsen stain was performed. Shigella spp., and Salmonella were also investigated by standard methods. An adequate stool dilution was cultivated on Petri plates with Mac Conkey agar. From each plate three lactose fermentative colonies were taken and standard biochemical tests were performed on them.
A total of 88 strains, biochemically identified as Escherichia coli, were analysed in the present study.
Serotyping
Identification of the somatic (O) antigens of the strains was done by the standard agglutination method, using monovalent serum provided by the Malbrán Institute ( Bs.As. , Argentina ). The investigated serogroups were the following: O26, O44, O55, O86, O111, O114, O119, O125, O126, O127, O128, O136, O142.
Tissue culture adhesion tests in HEp – 2 cells
Adhesion tests were performed in HEp–2 cells (ATCC: American Type Culture Collection), acquired from the Argentinean Cell Bank Association (ABAC). Cells were cultured in Minimum Essential Medium (MEM), with Earle salts (Sigma Co.), supplemented with 10% bovine fetal serum (Gen, Bs. As. , Argentina ), without antibiotics, in a stove at 37°C and 5% CO2. The method was adapted from Cravioto et al . in 1979. The HEp-2 cells were grown to 50-70% confluent monolayers on glass coverslips in tissue culture plates. After a gentle wash with phosphate-buffered saline (PBS), 1 mL of fresh MEM containing 2% fetal bovine serum and 0.5% D-mannose were added to each well, with 20 µL (2x105) of an overnight E.coli broth culture. The mixture was incubated for 3 h at 37°C in 5% CO2. After the incubation period, cells were washed several times with PBS, fixed with 70% methanol, stained with 10% Giemsa stain for 15 min and examined by light microscopy (100X). The reference strains used were: 2348/69: localized pattern, AA17-2 (O3:H2): aggregative pattern, RS 51-1: diffuse pattern, HS (O9:H4): negative adherence pattern. Samples were analyzed in duplicate to eliminate potential technical errors. Discordant pair results were repeated in duplicate, using the same technique.
Multiplex PCR
The E.coli strains were examined by Multiplex PCR to detect the genes ast A (which codify EAST1- Enteroaggregative Heat Stable Enterotoxin I) and agg R (Transcription Regulator for AAF/I and AAF/II). This reaction, which included primers for both genes, allowed the detection of E. coli that were bearing one or both characteristics.
For the PCR reaction, DNA was obtained from the sample in process. Bacteria were harvested from 1 mL of an overnight culture in TSB, suspended in 150 µL of Triton 1% on buffer TE, on an 1.5 mL-eppendorf tube. The tube was boiled for 10 min and centrifuged at 10,000 rpm. The supernatant was used as a template for the PCR reaction.
The primers for the amplification of a 108 bp of the astA gene were:
START 5´ GCC ATC ACA GTA TAT CCG 3´
STOP 5´ GCG AGT GAC GGC TTT GTA GT 3´
Nucleotide sequences corresponding to the fimbriae genes I and II from EAEC (AAF/I and AAF/II) were obtained. The expression of both fimbrial structures is regulated by a common gene, denominated aggR (transcription regulator gene). Thus, the design of specific primers for this gene, would allow to detect both types of fimbriae in only one reaction. The PC Gene Programm was used for the search of the right primers . From a variable number of selected primers by the program, one pair was chosen, which flanks a 430 nucleotides region:
START 5' AGA CGC CTA AAG GAT GCC C 3'
STOP 5' GAG TTA TCA AGC AAC AGC AAT GC 3'
The sequence of both primers was introduced on the GenBank data base in order to compare the sequence of all the incorporated genes. The result showed 100% homology with the aggR gene, access number Z32523, assuring that it would only amplify the desired gene.
The PCR mixture contained 10 mM Tris-HCl (pH 9 at 25°C ), 50 mM KCl and 0.1% Triton® X-100, MgCl2 1.5 mM , dNTP 0.2 mM each, 2 U of Taq polymerase (Promega), 1 µM each primer, water and 10 µL DNA.
The PCR program was: one step of 5 min at 94°C , 30 cycles of: 30 s at 94°C , 1 min at 57°C , 40 s at 72°C , one step of 5 min at 72°C .
AA17/2 (O3:H2) strain was used as the positive control, and HS (O9:H4) as the negative one. The strains were from the collection of the Center for Vaccine Development ( Maryland , USA ) (Fig. 4).
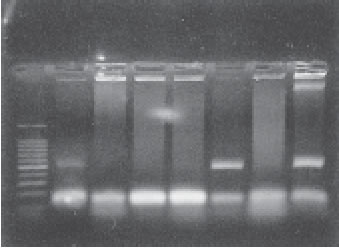
FIGURE 4. Agarose gel electrophoresis of PCR amplified DNA products. Lane 1 molecular size marker (100-bp ladder; Promega); lanes 3, 4, 5 and 7, astA PCR(108 bp); lanes 2, 6 and 7, astA and aggR PCR (108-bp and 430-bp respectively).
Statistical analysis
The sensitivity and specificity values were obtained by comparison between the HEp-2 assay and the PCR using Fisher exact test. A P value of < 0.05 was considered statistically significant.
Results
Based on the adhesion to HEp-2 cells, 34 strains (39%) showed the localized (LA) pattern; 23 strains (26%) were aggregative (AA) and 13 strains (15%) were diffuse. Eighteen strains (20%) did not show any adherence pattern. Figure 5 shows the results of the HEp-2 assay.
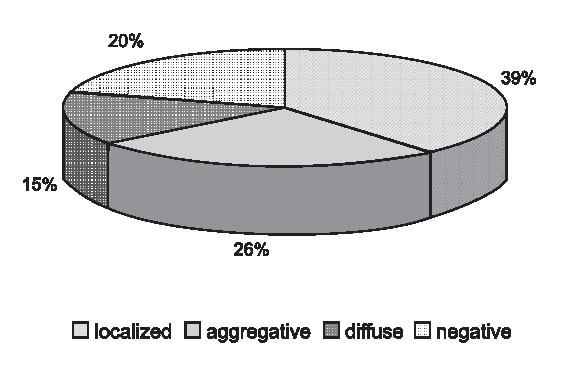
FIGURE 5. Distribution of adherence patterns.
Table 1 shows the results for aggR PCR and aggregative adherence pattern by HEp-2 test.
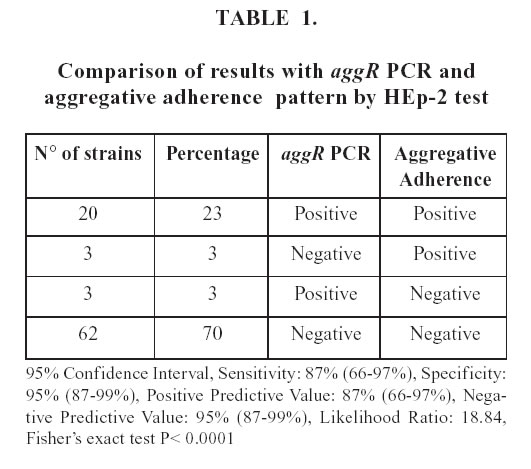
Twenty of the 23 aggregative pattern strains harbored the aggR gene, according to the PCR results, improving the sensitivity previously obtained.
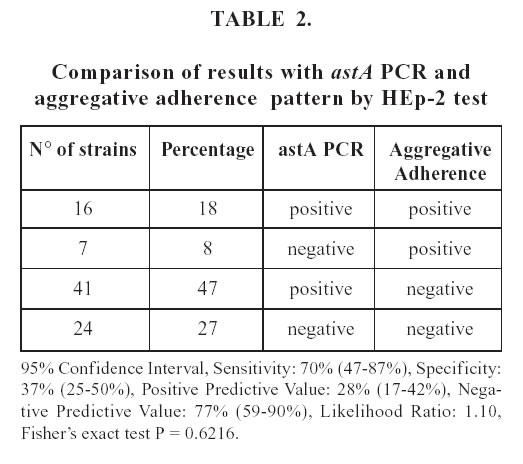
Table 2 compares the results of astA PCR and HEp-2 assay. Although 57 strains harbored the astA gene, only 16 of these expressed the AA pattern in the HEp-2 assay. Seven strains that presented the AA phenotype in the HEp-2 assay were negative for the astA gene. The association between both methods was not significant.
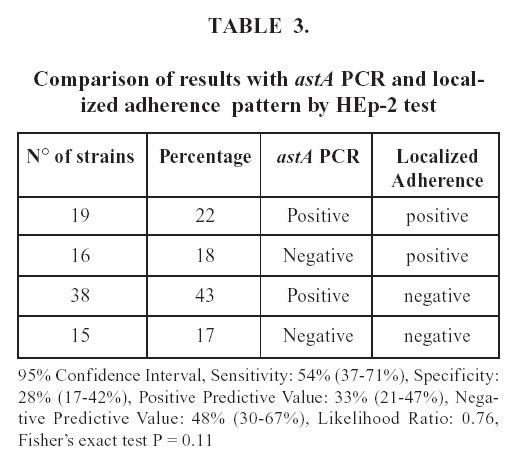
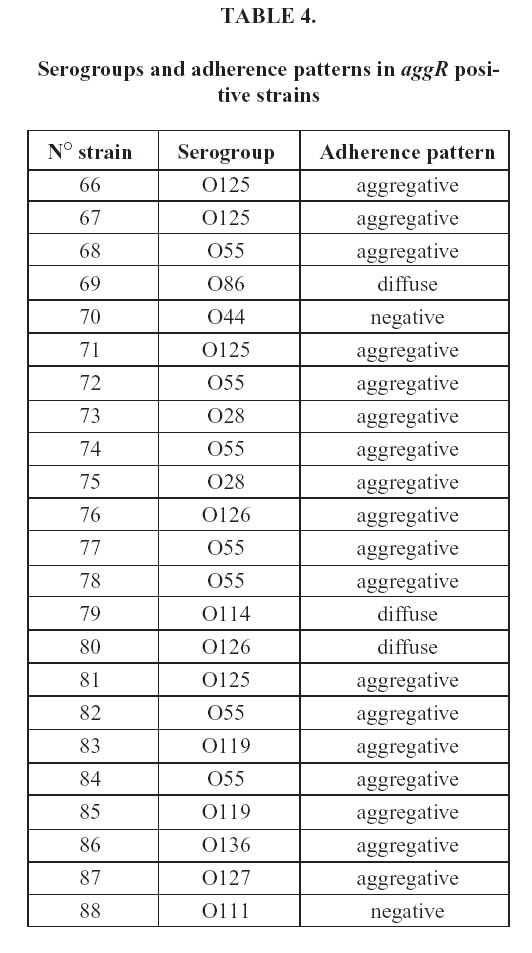
The astA gene have been detected not only in EAEC, but also in EPEC. Table 3 shows the comparison between this gene and the LA pattern, characteristic of EPEC. The astA gene was found in 57 (65%) of the strains, 19 (22%) of which showed the LA pattern. In addition 38 of the astA carrying strains presented another pattern of adherence. The serogroups and the adherence patterns of the aggR strains are described in Table 4. Among the serogroups found, O86, O44, O127 and O111 are epidemiologically related to EAEC; the rest belongs to Class I classical EPEC. The majority of aggR strains (78%) showed the aggregative pattern, three were diffuse and two negative.
Discussion
EAEC is a diarrheal pathogen of emerging importance, and it has been involved in both endemic pediatric diarrhea and outbreaks worldwide (Valentiner-Branth et al ., 2003; Dutta et al ., 1999). However, the pathogenic mechanisms of EAEC infection are not fully understood. Moreover, it appears to be a significant heterogeneity of virulence markers among EAEC isolates (Nataro et al ., 1995; Zamboni et al ., 2004). The specific role of the virulence determinants in the pathogenicity of diarrhea is being investigated. The demonstration of aggregative attachments of organisms to human epithelial cells (HEp-2) remains as the gold standard assay for EAEC. This test requires specialized facilities, and it can only be conducted in research laboratories (Miqdady et al ., 2002). The PCR offers new possibilities for the diagnosis because it is sensitive, specific, practical and has lower cost than the cell culture assay.
This study was designed to evaluate a multiplex PCR method for identification of EAEC. The virulence markers included in the PCR were aggR and astA , because both are widely distributed among EAEC strains (Nataro et al ., 1994; Yoshikazu et al ., 2002).
The distribution of the different adherence patterns varies between different reports (Cravioto et al .,1991; Nataro and Kaper, 1998; Okeke et al ., 2000; Ortiz et al ., 1998; Pabst et al ., 2003; Yatsuyanagi et al ., 2002). In this work, this distribution was similar to a previous study (Rüttler et al ., 2002), suggesting that the strains recovered from infected patients in our community are endemics. Studies to determine their epidemiologic and clonal relationship are being conducted in our laboratory.
Among the 23 strains adhering to HEp-2 cells with AA phenotype, twenty harbored sequences homologous to the aggR gene, according to the PCR results, demonstrating a high concordance between both methods. Other authors obtained similar results when examined the relationship between the possession of EAEC plasmid-borne gene aggR and the ability of E.coli strains to adhere to HEp-2 cells (Jiang et al ., 2002; Tsukamoto,1996).
Moreover, Tsukamoto (1996) proposed a PCR method based on the aggR gene; this method was shown to have greater sensitivity than the probe assay in detecting enteroaggregative Escherichia coli .
With respect to the astA gene, we did not find significant association with the aggregative phenotype in the HEp-2 assay. In fact, although 57 strains harbored the astA gene, only 16 of these expressed the AA pattern in the HEp-2 assay (Table 2).
The astA gene was initially identified in EAEC as a structural gene that encodes a distinct low-molecularweight putative enterotoxin (Savarino et al ., 1993). In the last years, the astA gene has been detected not only in EAEC but also in EPEC, atypical EPEC, ETEC, STEC and EIEC strains. We also compared the astA PCR with the LA pattern, characteristic from EPEC. An interesting finding of this work was that the astA gene had a similar distribution between aggregative pattern (18%) and localized pattern (22%) strains, indicating that this gene would not be considered as a marker of EAEC. Some studies proposed the denomination of EAST1EC (Enteroaggregative Stable Toxin Escherichia coli ) for the strains which presented astA gene as the only factor, because reports suggesting the diarrheagenicity of EAST1 gene-possessing E.coli have gradually increased (Nishikawa et al ., 2002; Zhou et al ., 2002; Yatsuyanagi et al ., 2002). Further studies would be necessary to clarify the importance of the astA gene as putative diarrheagenic factor.
The serogroups and the adherence patterns of the aggR strains are described in Table 4. Between the serogroups founded, O86, O44, O127 and O111 are epidemiologically related to EAEC, the rest belongs to Class I classical EPEC. According to this findings, the serogroup of E.coli is not likely to correspond to the pathogenic factors.
The sensitivity and specificity of aggR PCR were 78% and 95%, respectively. Compared with our previous results, the sensitivity was higher without modification of the specificity. The latter allows us to affirm that this method can detect more aggregative strains because the new gene is common to AAF/I and AAF/II strains.
Moreover, since in our study a strong correlation between the presence of the specific marker aggR and the reference test was found, we can conclude that aggR may be used to identify EAEC, using the PCR method as a screening test for the diagnosis of EAEC.
Since some strains that did not carry aggR gene adhered to HEp-2 cells with an aggregative pattern and could be pathogenics, the adhesion assay should be used to complement the diagnosis in the absence of other known pathogen or in persistent cases of diarrhea.
References
1. Baudry B, Savarino SJ, Vial P, Kaper JB, Levine MM (1990). A sensitive and specific DNA probe to identify enteroaggregative Escherichia coli , a recently discovered diarrheal pathogen. J Infect Dis. 161: 1249-1251. [ Links ]
2. Bhan MK, Raj P, Levine MM, Kaper JB, Bhandari N, Srivastava R, Kumar R, Sazawal S (1989). Enteroaggregative Escherichia coli associated with persistent diarrhea in a cohort of rural children in India . J Infect Dis. 159: 1061-1064. [ Links ]
3. Bhatnagar S, Bhan MK, Sommerfelt H, Sazawal S, Kumar R, Saini S (1993). Enteroaggregative Escherichia coli may be a new pathogen causing acute and persistent diarrhea. Scand J Infect Dis. 25: 579-583. [ Links ]
4. Cobeljic M, Miljkovic-Selimovic B, Paunovic-Todosijevic D, Velickovic Z, Lepsanovic Z, Savic D, Ilic R, Konstantinovic S, Jovanovic B, Kostic V (1996). Enteroaggregative Escherichia coli associated with an outbreak of diarrhoea in a neonatal nursery ward. Epidemiol Infect. 117: 11-16. [ Links ]
5. Cravioto A, Gross RJ, Scotland SM, Rowe B (1979). An adhesive factor found in Strains of Escherichia coli belonging to the traditional infantile Enteropathogenic Serotypes. Curr Microbiol. 3: 95-99. [ Links ]
6. Cravioto A, Tello A, Navarro A, Ruiz J, Villafan H, Uribe F, Eslava C (1991). Association of Escherichia coli HEp-2 adherence patterns with type and duration of diarrhoea. Lancet 337: 262-264. [ Links ]
7. Czeczulin JR, Balepur S, Hicks S, Phillips A, Hall R, Kothary M H, Navarro-García F, Nataro JP (1991). Aggregative Adherence Fimbria II, a second fimbrial antigen mediating aggregative adherence in Enteroaggregative Escherichia coli . Am Soc Microb. 65: 4135-4145. [ Links ]
8. Debroy C, Bright BD, Wilson RA, Yealy RD, Yumar R, Bhan MK (1994). Plasmid-coded DNA fragment developed as a specific gene probe for the identification of enteroaggregative Escherichia coli . J Med Microbiol. 41: 393-398. [ Links ]
9. Dutta S, Pal S, Chakrabarti S, Dutta P, Manna B (1999). Use of PCR to identify enteroaggregative Escherichia coli as an important cause of diarrhoea among children living in Calcutta , India . J Med Microbiol. 48(11): 1011-1016. [ Links ]
10. Elías WP, Suzart S, Trabulsi LR, Nataro JP, Gomes TA (1999). Distribution of aggA and aafA gene sequences among Escherichia coli isolates with genotypic or phenotypic characteristics, or both, of enteroaggregative Escherichia coli . J Med Microbiol. 48(6): 597-599. [ Links ]
11. Eslava C, Navarro-García F, Czeczulin JR, Henderson JR, Cravioto A, Nataro JP (1998). Pet, as autrotransporter enterotoxin from enteroaggregative Escherichia coli . Infect Immun. 63: 4949-4952. [ Links ]
12. Jiang Z, Greenberg D, Nataro JP, Steffen R, DuPont HL (2002). Rate of occurrence and pathogenic effect of enteroaggregative Escherichia coli virulence factors in international travelers. J Clin Microbiol. 40(11): 4185-4190. [ Links ]
13. Law D (1994). Adhesion and its role in the virulence of enteropathogenic Escherichia coli. Clin Microbiol Rev. 7: 152-173. [ Links ]
14. Miqdady MS, Jiang ZD, Nataro JP, DuPont HL (2002). Detection of enteroaggregative Escherichia coli with formalin-preserved HEp-2 cells. J Clin Microbiol. 40: 3066-3067. [ Links ]
15. Nataro JP, Kaper JB, Robins Browne R, Prado V, Vial P, Levine MM (1987). Patterns of adherence of diarrheagenic Escherichia coli to HEp-2 Cells. Pediatr Infect Dis J. 6: 829-831. [ Links ]
16. Nataro JP, Yikang D, Giron JA, Savarino SJ, Kothary MH, Hall R (1993). Aggregative adherence fimbria I expression in enteroaggregative Escherichia coli requires two unlinked plasmid regions. Infect Immun. 61: 1126-1131. [ Links ]
17. Nataro JP, Yikang D, Yingkang D, Walker K (1994). AggR, a transcriptional activator of aggregative adherence fimbria I expression in enteroaggregative Escherichia coli . J Bacteriol. 176: 4691-4699. [ Links ]
18. Nataro JP, Yikang D, Cookson S, Cravioto A, Savarino SJ, Guers LD, Levine MM, Tacket CO (1995). Heterogeneity of enteroaggregative Escherichia coli virulence demonstrated in volunteers. J Infect Dis. 171: 465-468. [ Links ]
19. Nataro JP, Kaper JB (1998). Diarrheagenic Escherichia coli . Clin Microbiol Rev. 11: 142-201. [ Links ]
20. Nataro JP, Steiner T, Guerrant RL (1998). Enteroaggregative Escherichia coli . Em Infect Dis. 4: 251-261. [ Links ]
21. Nataro JP, Cravioto A (1998). In vitro effects of a high-molecularweight heat labile enterotoxin from enteroaggregative Escherichia coli . Infect Immun. 66(7): 3149-3154. [ Links ]
22. Nishikawa Y, Zhou Z, Hase A, Ogasawara J, Kitase T, Abe N, Nakamura H, Wada T, Ishii E, Haruki K, Surveillance Team (2002). Diarrheagenic Escherichia coli isolated from stools of sporadic cases of diarrheal ilness in Osaka City , Japan between 1997 and 2000: Prevalence of Enteroaggregative E. coli Heat-Stable Enterotoxin 1 Gene-Possessing E.coli . Jpn J Infect Dis. 55(6): 183-190. [ Links ]
23. Okeke IN, Lamikanra A, Steinbruck M, Kaper JB (2000). Characterization of Escherichia coli strains from cases of childhood diarrhea in southwestern Nigeria . J Clin Microbiol. 38: 7-12. [ Links ]
24. Okeke IN, Lamikanra A, Czeczulin J, Dubovsky F, Kaper JB, Nataro JP (2000). Heterogeneus Virulence of enteroaggregative Escherichia coli strains isolates in Southwest Nigeria . J Infect Dis. 181: 252-60. [ Links ]
25. Okeke IN, Nataro JP (2001). Enteroaggregative Escherichia coli . The Lancet Inf Dis. 1: 304-13. [ Links ]
26. Ortiz A, Rüttler M, García B, Balbi L, Cruzado M, Risler N, Castro C (1998). Acumulación de actina y adherencia a células HEp-2 de cepas de Escherichia coli aisladas de niños con diarrea en Mendoza, Argentina. Rev Arg Microbiol. 30: 13-19. [ Links ]
27. Pabst WL, Altwegg M, Kind C, Mirjanic S, Hardegger D, Nadal D (2003). Prevalence of enteroaggregative Escherichia coli among children with and without diarrhea in Switzerland . J Clin Microbiol. 41(6): 2289-2293. [ Links ]
28. Piva IC, Pereira AL, Ferraz LR, Silva RSN, Vieira AL, Blanco JE, Blanco M, Blanco J, Giugliano LG (2003). Virulence markers of enteroaggregative Escherichia coli isolated from children and adults with diarrhea in Brasilia , Brazil . J Clin Microbiol. 41: 1827-1832. [ Links ]
29. Rich Ch, Favre-Bonte S, Sapena F, Joly B, Forestier Ch (1999). Characterization of enteroaggregative Escherichia coli Isolates. FEMS Microbiol Lett. 173: 55-61. [ Links ]
30. Rüttler ME, Renna NF, Balbi L, García B, Guidone L, Fernández R, Puig O, Ortiz A (2002). Characterization of enteroaggregative Escherichia coli strains isolated from children with acute diarrhea, in Mendoza , Argentina . Rev Arg Microbiol. 34: 167-170. [ Links ]
31. Savarino SJ, Fasano A, Watson J, Martin BM, Levine MM, Guandalini S, Guerry P (1993). Enteroaggregative Escherichia coli heat-stable enterotoxin 1 represents another subfamily of E. coli heat-stable toxin. Proc Natl Acad Sci USA 1;90(7): 3093-3097. [ Links ]
32. Schmidt H, Knop C, Franke S, Aleksic S, Heeseman J, Karch H (1995). Development of PCR for screening of enteroaggregative Escherichia coli . J Clin Microbiol. 33: 701-705. [ Links ]
33. Tsai CC, Chen SY, Tsen HY (2003). Screening the Enteroaggregative Escherichia coli activity and detection of the aggA, aafA, and astA genes with novel PCR primers for the Escherichia coli isolates from diarrhea cases in Taiwan . Diagn Microbiol Infect Dis. 46(3): 159-165. [ Links ]
34. Tsukamoto T (1996). PCR methods for detection of enteropathogenic Escherichia coli (localized adherence) and enteroaggregative Escherichia coli . Kansenshogaku Zasshi 70(6): 569-573 (Abstract). [ Links ]
35. Valentiner-Branth P, Steinsland H, Fischer TK, Perch M, Scheutz F, Dias F, Aaby P, Molbak K, Sommerfelt H (2003). Cohort study of guinean children: Incidence, pathogenicity, conferred protection, and atributable risk of Enteropathogens during de first 2 years of life. J Clin Microbiol. 41(9): 4238-4245. [ Links ]
36. Yatsuyanagi J, Saito S, Miyajima Y, Amano KJ, Enomoto K (2003). Characterization of atypical enteropathogenic Escherichia coli strains harboring the astA gene that were associated with a waterborne outbreak of diarrhea in Japan . J Clin Microbiol. 41: 2033-2039. [ Links ]
37. Yatsuyanagi J, Shioko S, Hiroyasu S, Yoshimichi M, Ken-Ichi A, Katsuhiko E (2002).Characterization of enteropatogenic and enteroaggregative Escherichia coli isolated from diarrheal outbreaks. J Clin Microbiol. 40(1): 294-297. [ Links ]
38. Yoshikazu N, Zhijiang Z, Atsushi H, Jun O, Teruyo K, Nichiro A, Hiromi N, Takayuki W, Eiji I, Kosuke H, and the Surveillance Team (2002). Diarrheagenic Escherichia Coli isolated from stools of sporadic cases of diarrheal illness in Osaka City , Japan between 1997 and 2000: Prevalence of enteroaggregative E. coli Heat -Stable Enterotoxin 1 gene- possessing E. coli . Jpn J Infect Dis. 55: 183-190. [ Links ]
39. Zamboni A, Fabbricotti SH, Fagundes-Neto U, Scaletsky ICA (2004). Enteroaggregative Escherichia coli Virulence Factors Are Found To Be Associated with Infantile Diarrhea in Brazil . J Clin Microbiol 42(3): 1058-1063. [ Links ]
40. Zhou Z, Ogasawara J, Nishikawa Y, Seto Y, Helander A, Hase A, Iritani N, Nakamura H, Arikawa K, Kai A, Kamata Y, Hoshi H, Haruki K (2002). An outbreak of gastroenteritis in Osaka , Japan due to Escherichia coli serogroup O166:H15 that had a coding gene for enteroaggregative E. coli Heat-Stable Enterotoxin 1 (EAST1). Epidemiol Infect. 128(3): 363-371. [ Links ]
Received on December 15, 2005.
Accepted on March 16, 2006.














