Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
Biocell
versión impresa ISSN 0327-9545
Biocell v.30 n.3 Mendoza ago./dic. 2006
Cysteine proteinases of Trypanosoma cruzi: from digestive enzymes to programmed cell death mediators
Gregor Kosec*¶, Vanina Alvarez**¶ and Juan J. Cazzulo**.
* Jozef Stefan Institute, Jamova 39, SI 1000 Ljubljana, Slovenia.
** Instituto de Investigaciones Biotecnológicas (IIB-INTECH), Universidad Nacional de General San Martín - CONICET. Av. Gral. Paz 5445, (1650) San Martín, Buenos Aires, Argentina.
¶ These authors contributed equally to the present work. Address correspondence to: Dr. Juan José Cazzulo. Instituto de Investigaciones Biotecnológicas (IIB-INTECH). Universidad Nacional de General San Martín - CONICET. Av. Gral. Paz 5445, Casilla de Correo 30, (1650) San Martín, Buenos Aires, ARGENTINA. E-mail: jcazzulo@iib.unsam.edu.ar Fax: (+54-11) 4752 9639
ABSTRACT: Trypanosoma cruzi, the parasite causing Chagas disease, contains a number of proteolytic enzymes. The recent completion of the genome sequence of the T. cruzi CL Brener clone suggests the presence of 70 cysteine peptidases, 40 serine peptidases (none of them from the chymotrypsin family), about 250 metallopeptidases (most leishmanolysin homologues), 25 threonine peptidases, and only two aspartyl peptidases, none of them from the pepsin family. The cysteine peptidases belong to 7 families of Clan CA, 3 families of Clan CD, and one each of Clans CE and CF. In Clan CA, the C1 family is represented by cruzipains 1 and 2, biochemically well characterized, as well as cathepsin B and two other cathepsins. There are a number of homologues to calpains (family C2), probably non-functional, lacking the Ca-binding domain. Family C54 includes the Atg4 proteinases (autophagins), which seem to be involved in the autophagic process. Clan CD includes family C14, the metacaspases. We have expressed the metacaspases TcMCA3 and TcMCA5, and obtained indirect evidence of their participation in programmed cell death induced by fresh human serum in the parasite. More experiments are required to better define their role in apoptosis.
Key words: Trypanosoma cruzi; Chagas disease; Cysteine proteinases; Apoptosis; Autophagy
Introduction
Trypanosoma cruzi, a flagellated protozoan parasite, is the causative agent of the American trypanosomiasis, Chagas disease. The parasite has a complex life cycle, involving two forms present in the gut of the insect vector, the replicative epimastigote and the infective metacyclic trypomastigote, and two forms present in the infected mammal, the intracellular amastigote and the bloodstream trypomastigote released from infected cells into the blood. This infection is endemic in Latin America; its prevalence is estimated at 16-18 million cases, with about 120 million people at risk. No vaccines have been developed so far; the low effectiveness of the chemotherapeutic agent available (at present, only benznidazole) together with its undesirable side effects, makes treatment of Chagas disease difficult. There is, therefore, an urgent need for the identification of novel drug targets to improve the treatment of this disease (Barrett et al., 2003).
The study of peptidases in protozoan parasites in general, and in Trypanosomatids in particular, has acquired considerable importance over the last few years. Some peptidases have been proposed to play central roles in diverse processes such as cell invasion, differentiation, cell cycle progression, catabolism of host proteins and evasion of the host immune response (Klemba and Goldberg, 2002). The possibility of developing selective inhibitors of key proteinases of parasites is being explored as a novel chemotherapeutic strategy.
T. cruzi has been shown to contain several proteolytic activities (Cazzulo, 2002), which have been biochemically characterized; other peptidases have been predicted from the data of the recently completed genome project (El-Sayed et al., 2005). Although the T. cruzi genome still presents a number of uncertainties, predictions so far (Ivens et al., 2005, Tables S17 and S18) include about 70 putative cysteine peptidases, about 40 putative serine peptidases (none of them belonging to the S1 family), about 250 putative metallopeptidases, of which most are homologues of leishmanolysin, 25 putative threonine peptidases, most of them homologues of proteasome subunits, and 2 putative aspartyl peptidases. It is noteworthy that among the latter, the complete absence of A1 family peptidases (pepsin-like) has been reported for the genomes of the three trypanosomatids, the only genes detected encoding homologues to presenilin 1 and signal peptide peptidase. The actual total number of proteolytic enzymes in T. cruzi is probably about a half, since in a number of cases the alleles have been reported as different genes; it is likely that the total number, once properly determined, will be closer to the numbers proposed for both Trypanosoma brucei and Leishmania major, about 160 in both cases. Among the enzymes biochemically characterized, we can mention cysteine proteinases, such as cruzipain (Cazzulo, 2002), a 30-kDa cathepsin B-like cysteine proteinase (Garcia et al., 1998), and the recently described metacaspases (Kosec et al., 2006); two serine proteinases belonging to the prolyl oligopeptidase family (Burleigh et al., 1997; Bastos et al., 2005) and a serine carboxypeptidase (Parussini et al., 2003); a glycosylphosphatidylinositol (GPI)-anchored membrane metalloproteinase of the leishmanolysin family (Cuevas et al., 2003), and the proteasome (de Diego et al., 2001; Gonzalez et al., 1996).
Apoptosis or programmed cell death (PCD) is a physiological process that is of central importance for the development and homeostasis of multicellular organisms; it is mediated by a cascade of cysteine proteinases, the caspases (Grutter, 2000). Phenomena resembling apoptosis have been described in a variety of plants and unicellular eukaryotes, which apparently lack direct caspase homologues, and this was accompanied by an increase in caspase-like proteolytic activity (cleavage of substrates after an Asp residue) (Madeo et al., 2002; Das et al., 2001; Bozhkov et al., 2004). A process resembling apoptosis has been described in T. cruzi, and can be induced by different stimuli, including treatment of epimastigotes with fresh human serum (FHS) (Ameisen, 1995; Piacenza et al., 2001). This process could be of great importance enabling the parasite to avoid an early inflammatory response if epimastigotes enter the mammalian bloodstream (Zeledon et al., 1984) or could render host macrophages more susceptible to invasion as shown for apoptotic T cells (Freire-de-Lima et al., 2000). With the discovery of metacaspases, distant caspase-homologues (Uren et al., 2000) in genomes of plants and unicellular eukaryotes, their role in PCD became an intriguing possibility. We have recently shown that this is probably the case in T. cruzi (Kosec et al., 2006).
We describe here an analysis of the cysteine peptidase families and enzymes predicted from the T. cruzi genome sequence data, as well as a further characterization of proteins belonging to family C14 (metacaspases), and their possible role in apoptosis.
Materials and methods
Genomic database searches
Aminoacid sequences corresponding to the catalytic domains of characteristic members of the known cysteine peptidase families were obtained in the MEROPS peptidase database (http://merops.sanger.ac.uk) (Rawlings et al., 2006). These sequences were entered as query in the tBLASTn searches on the predicted genes databases of T. cruzi at the GeneDB web page (www.genedb.org). The hits with sufficient similarity were analyzed for the presence of the predicted catalytic residues.
Parasite cultures
T. cruzi epimastigotes of the CL Brener clone (Zingales et al., 1997) were cultured in brain-hearttryptose (BHT) medium (Franke de Cazzulo et al., 1994). For induction of apoptosis, epimastigotes were grown to a density of 5 x 107/ml. Fresh human serum (FHS) was added to 10% final concentration and cultures were incubated for 4 h at 28ºC. The proportion of apoptotic cells was estimated by morphology and motility, as previously described (Kosec et al., 2006).
Enzymatic assays
Epimastigotes were washed and resuspended in PBS (10 mM Na2HPO4, 150 mM NaCl, pH 7.2) and cells were ruptured by three rounds of freezing at -20ºC and thawing. The cell-free extract was obtained by centrifugation at 26,900 x g for 15 min at 4ºC and the pellet was discarded. The fluorogenic substrate Z-YVAD AFC (Biomol) was used to measure caspase-like activity. Samples were preincubated in caspase buffer (50 mM HEPES pH 7.5, 100 mM NaCl, 0.1% CHAPS, 10% sucrose, 1 mM EDTA, 20 mM DTT) in the presence of 100 mM E-64 for 10 min at room temperature. Afterwards, 100 mM Z-YVAD-AFC was added and changes in fluorescence emission were followed for 30 min at 25°C. The same cell free extracts were used to measure cruzipain activity. Samples were preincubated in 50 mM Tris, 10 mM β-mercaptoethanol for 10 minutes at room temperature. After that the colorimetric assays were performed with 150 mM Bz-PFR-pNA (Sigma) and absorbance was measured at 410 nm.
Immunofluorescence microscopy
Epimastigotes were left to bind to poly-L-lysinecoated slides, fixed with 4% (w/v) paraformaldehyde in PBS and washed twice. Blocking buffer (3% goat serum, 2% bovine serum albumin, 0.1% saponine in PBS) was used for the saturation of coverslides. Polyclonal antibodies against metacaspase-3 (TcMCA3) and metacaspase-5 (TcMCA5) (Kosec et al., 2006) were then diluted 1/250 in blocking buffer and used for incubation of the fixed parasites. Coverslides were washed three times and incubated with AlexaFluor 488-conjugated goat anti-mouse immunoglobulins (Molecular Probes) for 1 h at room temperature. After extensive washing, DNA was stained with 5 mg/ml 4',6-diamidino- 2-phenylindole (DAPI) and cover slips were mounted using FluorSaveTM Reagent (Calbiochem). Preparations were analyzed using a fluorescence microscope (Nikon Eclipse E600) and image capture was performed by using a Spot RT Slider Model N¼ 2.3.1 digital camera (Diagnostic Instruments).
In-vivo determination of active caspases with SR-VADFMK fluorescent probe
Epimastigote cultures undergoing apoptosis were stained for caspase activity using SR-VAD-FMK fluorescent probe (Sulforhodamine Multi-Caspase Activity Kit Apoptosis Detection, Biomol) following the manufacturerÕs instructions. Cells were then attached to poly-L-lysine-coated glass slides and processed for fluorescence microscopy observation as described above.
Overexpression of metacaspases in T. cruzi epimastigotes
The complete ORFs of TcMCA3 and TcMCA5 (Kosec et al., 2006) were cloned into the pRibotex plasmid (Martinez-Calvillo et al., 1997). The constructions, together with an empty pRibotex as control, were electroporated into epimastigotes. Geneticin (G418 sulphate)(Life Technologies) was used as selection marker as previously described (Kosec et al., 2006).
Results and Discussion
Cysteine peptidases in Trypanosoma cruzi
The recently completed genome of T. cruzi allowed a preliminary survey of the cysteine peptidases present in the parasite. Seven families of Clan CA are potentially present. The C1 family, the prototype of which is papain, contains the well characterized cruzipains (cruzipain 1 and 2) and cathepsin B (Cazzulo, 2002) and, in addition, two other putative members annotated as cathepsin S-like and bromelain-like proteins. Cruzipain is a member of a large family composed of polymorphic genes. The majority of them encodes highly similar isoforms (approximately 98% amino acid identity) and were termed as cruzipain 1, while the most divergent cruzipain gene (approximately 88% identical to cruzipain 1) was termed cruzipain 2. Genome data from the T. cruzi CL Brener clone predicts the presence of a number of cruzipain 1 genes; however, the high degree of sequence identity among the corresponding Genome Survey Sequences (GSS) makes unfeasible their exact discrimination into individual genes. Cathepsin B and cathepsin S seem to be present as a single copy per haploid genome since two alleles (sharing 96% and 95% amino acid identity, respectively) were found for each protein. Finally, one sequence homologous to bromelain was detected in the parasite genome. Figure 1 compares the sequences of one representative cruzipain 1 isoform, cruzipain 2 and the cathepsin B alleles with the newly identified cathepsin S alleles and the bromelain-like protein. The conserved C-H-N catalytic triad is indicated (*). Clan CA peptidases are characterized by having substrate specificity defined by the S2 pocket. In the case of cruzipains, a Glu residue (indicated by an arrowhead) has been shown to determine the substrate specificity for this enzyme, conferring the capacity to accept both hydrophobic and basic aminoacid residues at the S2 subsite. In addition, structural data from cruzipain 1 has pointed out the relevance of three amino acid residues (also indicated by arrowheads) in mediating substrate interaction within the S2 pocket; two replacements found in cruzipain 2 are thought to explain some differences in substrate specificity among these enzymes (dos Reis et al., 2006). Members of the CA clan are normally targeted to intracellular vesicle compartments or secreted; thus, they possess a leader peptide. In fact, this is the case for the T. cruzi enzymes as all the sequences present a predictable signal peptide at their N-terminus. In addition to the putative signal peptide, TMHMM2.0 program (www.genedb.org) predicts a transmembrane domain in the bromelain-like protein, suggesting that this protein might be membrane-anchored. It is noteworthy that the putative cathepsin S is annotated as presenting an Nterminal extension as compared with the other proteins; however, the existence of a second in frame Met residue well aligned with the initial Met of the cruzipains and the putative bromelain, as well as the relative position of the putative signal peptide, suggest that the second one is really the initial Met. Given the number of amino acid substitutions among the T. cruzi C1 peptidases, inside the catalytic domain and in particular within the S2 subsite, we might speculate that they represent different enzymes playing diverse roles in the parasite. Only when these new enzymes are expressed, we shall be able to know if they are actually active proteinases and to try to gain insights on their possible functions.
FIGURE 1. Alignment of predicted sequences of T. cruzi Family C1 cysteine proteinases. The sequences correspond to cruzipain-11 (AAA30181), cruzipain-21 (AAC37213), the cathepsin B alleles (1: Tc00.1047053511827.1002 and 2: Tc00.1047053510535.1002), cathepsin S alleles (1: Tc00.1047053507849.502 and 2: Tc00.1047053508317.102) and the bromelain-like sequence Tc00.1047053508173.1102. (1) denotes the GenBank accession number and (2) denotes the systematic name in GeneDB (www.genedb.org). Asterisks indicate the residues forming the conserved catalytic triad, and arrow heads indicate residues involved in the S2 pocket.
Family C2 (calpains), well characterized in mammals, includes heterodimeric proteins, consisting of a large subunit and a small subunit; the former contains four domains, among which domain II is the proteinase catalytic domain and domain IV contains calcium-binding motifs. The genomes of T. cruzi, T. brucei and L. major contain several genes each, encoding putative proteins belonging to this family. Some of them consist exclusively of domain I, of unknown function, and have been termed SKCRPs (small kinetoplastid calpain-related proteins), and some of them contain a typical domain II and were termed CALPs (calpain-like proteins) (Ersfeld et al., 2005). The T. cruzi CL Brener clone genome contains 24 sequences related to calpains, 15 CALPs and 9 SKRCPs. However, only two of the CALPs have an intact catalytic triad, and none has a domain IV, nor five amino acid residues in domain II which are also involved in calcium binding. Therefore, these data make rather unlikely that these proteins are active calpains; none of them has been expressed, and so their possible role as proteinases is not proved. It is noteworthy, however, that these proteins are conserved in the three trypanosomatids studied, and we may assume, therefore, that they are likely to play some function, so far unknown.
Ubiquitin and ubiquitin-like proteins are conjugated to many intracellular proteins, acting in the regulation of a number of cellular processes, including proteasomal protein degradation. The proteasome binds proteins linked through a Lys residue to four or more ubiquitin molecules, connected through their Lys48 residue. Before proteolysis, ubiquitin is deconjugated by deubiquitinating enzymes. The cysteine peptidase families C12, C19 and C65 consist of these enzymes. Family C12 has ubiquitin Cterminal hydrolases, which cleave different substrates, among them newly synthesized ubiquitin precursors. Two members of this family, homologous to two of the four ubiquitin C-terminal hydrolases encoded by the human genome, have been identified in the T. cruzi genome. Proteomic studies (www.tcruzidb.org) have shown that they are expressed at least in some developmental stages of the parasite. The C19 family consists of ubiquitin-specific proteinases, which represent the majority of the deubiquitinating enzymes present in the human genome, with some specific functions, although our knowledge of them is still far from complete. The T. cruzi genome encodes a group of 18 considerably divergent homologues of these proteins. The C65 family, which includes human otubain-1 and -2, with functions still not completely understood, is represented in T. cruzi by an otubain-1 homolog (Nijman et al., 2005; Amerik and Hochstrasser, 2004; Balakirev et al., 2003).
The C51 family consists of proteins having the CHAP (cysteine, histidine-dependent amidohydrolases/ peptidases) domain. Many proteins containing this do main are involved in cell wall metabolism in bacteria. This domain has also been found in proteins from bacteriophages and archea, and trypanosomatids among eukaryotes (Rawlings et al., 2006). The T. cruzi genome contains five sequences annotated as putative D-alanylglicyl endopeptidases. Their possible expresion and function in the parasite is unknown.
Family C54 includes the Atg4 proteinases, named autophagins because of their participation in the autophagic process by cleaving the Atg8 ubiquitin-like protein, which is involved in the formation of the autophagic vesicles (Marino et al., 2003). Two homologous proteins, TcAtg4.1 and TcAtg4.2, are present in the T. cruzi genome (Fig. 2); recently, we have been able to express them as well as their substrates TcAtg8.1 and TcAtg8.2. Both autophagins are able to cleave the TcAtg8 proteins at the conserved Gly residue, which then becomes linked to a phospholipid for membrane insertion (Ichimura et al., 2000). The expression of autophagin activity in the four major developmental stages of the parasite was shown by incubation of cellfree extracts with the recombinant TcAtg8 proteins (V. Alvarez et al., unpublished results).
FIGURE 2. Alignment of protein sequences of the predicted catalytic domains of autophagins. Atg4.1 and Atg4.2 genes are included from the trypanosomatid species (Tc - Trypanosoma cruzi - DQ 7682971 and DQ7682981; Tb: Trypanosoma brucei - Tb927.6.16902 and Tb11.01.79702; Lm - Leishmania major - LmjF32.38902 and LmjF30.02702). Yeast Atg4 (Sc - Saccharomyces cerevisiae - NP 0141761) and the most relevant human enzyme Atg4B (Hs - Homo sapiens - NP 0374571) are also aligned. Asterisks denote the catalytic site C, D and H residues as derived from the HsAtg4B crystal structure and a triangle marks the position of the Y residue, involved in the oxyanion hole formation. (1) denotes the GenBank accession number and (2) denotes the systematic name in GeneDB (www.genedb.org).
Clan CD is represented in the parasite genome by families C13, C14 and C50. Family C13 includes the GPI transamidase, involved in the transfer of the glycosyl- phosphatidyl inositol moiety to the proteins to be linked to the membrane by a GPI anchor (Mottram et al., 2003). GPI synthesis is highly relevant for trypanosomatids, since, contrary to mammals, most of their membrane proteins are linked through GPI anchors. One homologous protein is present in the T. cruzi CL Brener genome. Since the homologous proteins from T. brucei (Lillico et al., 2003) and L. mexicana (Ellis et al., 2002) have been shown to be active enzymes, it is very likely that the T. cruzi GPI8 is also an active protein. Figure 3 compares the respective sequences.
FIGURE 3. Alignment of predicted sequences of the GPI8 enzymes from the three trypanosomatids. Tc - Trypanosoma cruzi- Tc00.1047053511277.4502; Tb - Trypanosoma brucei - CAD291141; Lm - Leishmania major - CAJ035441. (1) denotes the GenBank accession number and (2) denotes the systematic name in GeneDB (www.genedb.org). Asterisks denote the catalytic H and C residues.
Family C14 includes the metacaspases; two different enzymes of this family are encoded in the T. cruzi genome. Figure 4 compares the amino acid residues spanning the active site of TcMCA3 and TcMCA5 with those of the enzymes from other Trypanosomatids, yeast and human caspase 3. Some experiments exploring their possible participation in apoptosis in epimastigotes will be described in the next section.
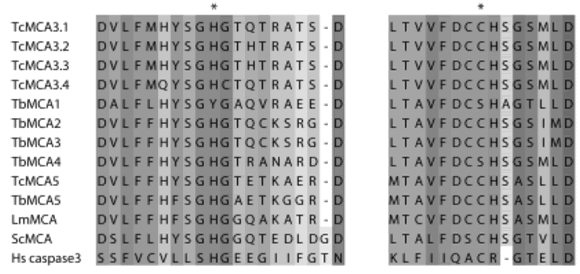
FIGURE 4. Partial amino acid sequences spanning the catalytic residues (*) are shown in the alignment of metacaspases from T. cruzi: TcMCA3.1 (AAY845801), TcMCA3.2 (AAY845811), TcMCA3.3 (AAY845821), TcMCA3.4 (AAY845831), TcMCA5 (AAY845791); T. brucei: TbMCA1 (Tb 11.02.07302), TbMCA2 (Tb 927.6.9402), TbMCA3 (Tb 927.6.9302), TbMCA4 (Tb 10.70.52502), TbMCA5 (Tb 09.211.47602); L. major: LmMCA (LmjF35.15802); Saccharomyces cerevisiae ScMCA (AAT928511) and human caspase-3 (Hs caspase-3, NP0043371). (1) denotes the GenBank accession number and (2) denotes the systematic name in GeneDB (www.genedb.org). Asterisks denote the catalytic H and C residues.
Family C50 consists of the separases, proteinases involved in the cleavage of the cohesin complex which maintains linked the sister chromatids during mitosis (Uhlmann, 2001). The separase must act only during the transition from metaphase to anaphase, and to this purpose it is kept in an inactive state as a complex with securin, which is degraded to allow the separase proteolytic action. Separases are big proteins, which have the proteinase domain at the C-terminus. The genome of T. cruzi contains two genes homologous to separases, most likely alleles since they have 98% identity, encoding proteins of about 1,100 amino acid residues.
Clan CE is represented in T. cruzi by one member of the family C48, of the deSUMOylating enzymes. In addition to ubiquitin, there are several ubiquitin-like proteins, which are conjugated to proteins with different purposes. One of them is the protein SUMO (Nacerddine et al., 2005). The deSUMOylating enzymes are involved in the deconjugation of the SUMO-protein complexes, which are likely to occur in the parasite since a SUMO homologue is present in the T. cruzi genome.
Finally, Clan CF is represented by family C15, of the pyroglutamyl peptidase I. This enzyme, like the M1 metallopeptidase pyroglutamyl peptidase II, cleaves the pyroglutamyl residue at the N-terminus of posttranslationally modified proteins, like some neuropeptide hormones (for instance, TRH, thyrotrophin-releasing hormone). One member of the C15 family is present in the T. cruzi genome. This is very interesting, since the homologous enzyme from T. brucei has been recently purified from the plasma of infected rats, which compared with the plasma of non-infected animals shows a much shorter half-life of TRH, and also of the gonadotropin- releasing hormone, GnRH, both in vitro and in vivo (Morty et al., 2006). These authors proposed the enzyme as a new virulence factor in sleeping sickness, participating in the endocrine lesions which are observed in African trypanosomiasis. The T. cruzi putative peptidase presents a high degree of sequence identity with the T. brucei enzyme (Fig. 5). If it is expressed by the mammalian stages of the parasite, the pyroglutamyl peptidase might also be important in the pathogenesis of Chagas disease.
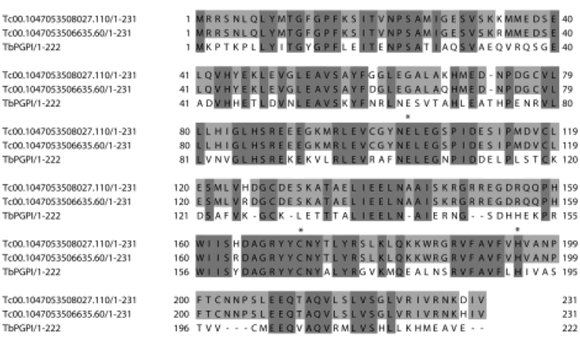
FIGURE 5. Alignment of the sequences of the pyroglutamyl peptidases I from Trypanosoma cruzi (Tc00.1047053508027.1102 and Tc00.1047053506635.602) and Trypanosoma brucei (AAY402941). (1) denotes the GenBank accession number and (2) denotes the systematic name in GeneDB (www.genedb.org). Asterisks denote the catalytic E, C and H residues.
Metacaspases and apoptosis in Trypanosoma cruzi
Proteolytic activities in cell-free extracts during FHS induced programmed cell death
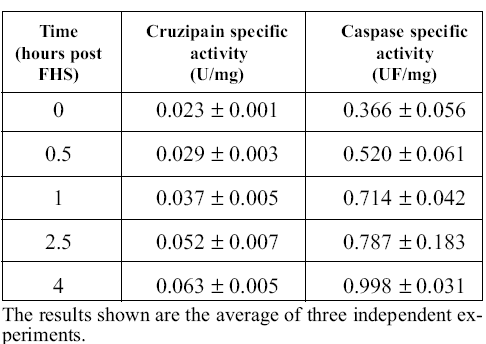
To elucidate whether proteolytic activities are involved in PCD of epimastigotes, we performed colori metric and fluorogenic measurements of cruzipain and caspase-like activities in the cell-free extracts obtained at different times after FHS exposure. The process usually takes about 4 hours to complete if 10% FHS is added to the culture. We observed the expected morphological changes in the course of the experiment (Kosec et al., 2006). Table 1 shows that both activities increased clearly during the process. Caspase-like activity measurements were performed in the presence of E-64, a potent irreversible cruzipain inhibitor to avoid interference from this very abundant and broadly specific protease, which is able to slowly cleave caspase substrates (specific activity 8.9 nmoles/min/mg of protein with the substrate DEVD-pNA, 0.3 mM, at 25ºC; this activity was insensitive to the caspase inhibitor DEVD-CHO at 0.2 mM, but was completely inhibited by E-64 at 20 mM). We have no simple explanation for the increase in cruzipain activity during apoptosis; it may be related to an increased necessity of hydrolytic activities during PCD to help cellular disorganization.
TABLE 1. Cruzipain and caspase activities after FHS treatment.
Subcellular localization of proteases during FHS induced programmed cell death
To discern in more detail the possible role of cruzipain and metacaspases in PCD we also performed immunofluorescence studies to observe their subcellular localization. In addition, a fluorescent activitybased probe for caspases was used to possibly connect the observed increased caspase-like activity with metacaspase localization. Figure 6 shows a remarkable change in the localization of metacaspases upon induction of apoptosis. Both metacaspases appear to be localized all over the cell body in normal conditions; however, they seem to colocalize with DAPI staining and thus, concentrate in the degrading nucleus after exposure to FHS. Interestingly, the SR-VADFMK signal, that specifically appears during the death process, also seems to partially colocalize with DAPI staining (Kosec et al., 2006). A similar finding was recently published for a plant metacaspase which also translocates from the cytosol to the nucleus, colocalizes with the nuclear pore complex and degrades the nuclear envelope (Bozhkov et al., 2005). On the other hand, Figure 6 also shows that cruzipain remains localized to the lysosomes/reservosomes during the whole process although these compartments seem to grow substantially as a result of FHS treatment, as compared with control epimastigotes (Fig. 6C). This further confirms that the observed caspase-like activity is cruzipain-independent.
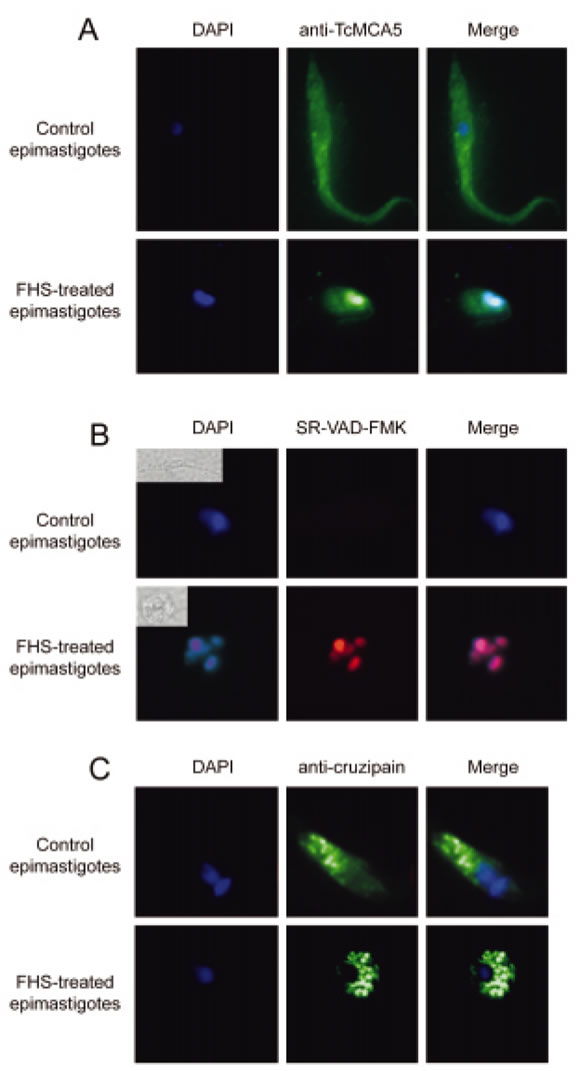
FIGURE 6. A) Fluorescence microscopy of DAPI-staining (left), anti-TcMCA5 antibody signal (centre) and a merged image (right) of FHS-treated epimastigotes (lower row) and control epimastigotes (upper row). FHS treatment leads to a change in localization of TcMCA5, which becomes colocalized with the degrading nucleus. B) Equivalent analysis of FHS-treated (lower row) and untreated (upper row) epimastigotes with DAPI and SR-VAD-FMK, fluorescent caspase-activity probe. SR-VAD-FMK signal is absent in control parasites and clearly colocalizes with DAPI staining after FHS treatment (lower right). Bright field images of the analysed parasites are shown in upper left corners in the right column. C) Localization of DAPI-staining (left), anticruzipain antibody signal (centre) and a merged image (right) in control (upper row) and FHS-treated (lower row) epimastigotes. Cruzipain clearly remains localized in the reserovosomes during the whole process.
Since the metacaspases behave differently than classical caspases during PCD, we were also interested in the processing of metacaspases during apoptosis. Caspases are synthesized as inactive single chain proenzymes and become active after apoptosis induction. Activation is accompanied by several cleavages of their polypeptide chain usually yielding two fragments of about 20 kDa and 10 kDa corresponding to the large and small subunits, respectively (Grutter, 2000). Nevertheless, no such processing could be observed for theTcMCA3 and TcMCA5 at any time of the death process (not shown). Together these results suggest that, although metacaspases are likely to be involved in PCD of T. cruzi, the mechanism seems to be clearly distinct from that of the metazoan caspases.
Caspase-like proteolytic activities in metacaspasetransfectant epimastigotes
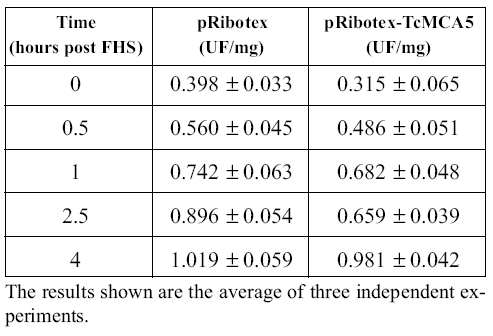
To clarify the connection between the metacaspases and caspase-like activity, we transfected epimastigotes with the pRibotex plasmid expressing the complete ORFs of TcMCA3 and TcMCA5. Transfections with TcMCA3 were unsuccessful, suggesting the possibility that this protein is lethal when overexpressed in the parasite. On the other hand, TcMCA5 transfectants were readily selected and grown in cultures without any special phenotype. However, they were more prone to undergo FHS-induced PCD, showing higher proportion of dead epimastigotes at all intermediate time points as compared with the control strain, transfected with an empty pRibotex vector (Kosec et al., 2006). We were also interested to see whether there would be higher caspase-like activity in cell-free extracts of TcMCA5- overexpressing parasites. As shown in Table 2, YVADase activity was very similar in both strains suggesting that metacaspases are not directly responsible for the augmented caspase-like activity. Possibly, metacaspases must activate other proteases that are responsible for this activity. Interestingly, such cell death specific proteases were recently isolated from plants and belong to the completely unrelated subtilisin family of serine-peptidases (Coffeen and Wolpert, 2004). It is remarkable that the genome of T. cruzi, CL Brener clone, contains four sequences, probably corresponding to the alleles of two different genes, annotated as putative subtilisinlike enzymes. Plant metacaspases were reported to specifically cleave substrates after Arg or Lys residues, rather than Asp, like the classical caspases (Vercammen et al., 2004). Unfortunately, we have not been able to detect proteolytic activity in the T. cruzi recombinant enzymes. Therefore, the specificity of trypanosomatid metacaspases remains unknown as well as their exact role in PCD. Further experiments are clearly necessary to elucidate the characteristics of these novel enzymes.
TABLE 2. Caspase activity in cell-free extracts of epimastigotes transfectants after FHS treatment.
Acknowledgements
This work was aided by grants from ANPCyT, SECYT, Argentina (PICT-01-06578) and the binational Argentinian-Slovenian Cooperation Agreement (ES/ PA02/S01). V.A. is a research fellow, and J.J.C. a member of the Research Career, from the Argentinian National Research Council (CONICET).
References
1. Ameisen JC, Izidorek T, Billaut-Mulot O, et al. (1995). Apoptosis in a unicellular eukaryote (Trypanosoma cruzi): implications for the evolutionary origin and role of programmed cell death in the control of cell proliferation, differentiation and survival. Cell Death Differ. 2: 285-300. [ Links ]
2. Amerik AY, Hochstrasser M (2004). Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 1695: 189-207. [ Links ]
3. Balakirev MY, Tcherniuk SO, Jaquinod M, Chroboczek J (2003). Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 4:517-522. [ Links ]
4. Barrett MP, Burchmore RJ, Stich A, Lazzari JO, Frasch AC, Cazzulo JJ, Krishna S (2003). The trypanosomiases. Lancet. 362: 1469-1480. [ Links ]
5. Bastos IM, Grellier P, Martins NF, Cadavid-Restrepo G, de Souza- Ault MR, Augustyns K, Teixeira AR, Schrevel J, Maigret B, da Silveira JF, Santana JM (2005). Molecular, functional and structural properties of the prolyl oligopeptidase of Trypanosoma cruzi (POP Tc80), which is required for parasite entry into mammalian cells. Biochem J. 388: 29-38. [ Links ]
6. Bozhkov PV, Filonova LH, Suarez MF, Helmersson A, Smertenko AP, Zhivotovsky B, von Arnold S (2004). VEIDase is a principal caspase-like activity involved in plant programmed cell death and essential for embryonic pattern formation. Cell Death Differ. 11: 175-182. [ Links ]
7. Bozhkov PV, Suarez MF, Filonova LH, Daniel G, Zamyatnin AA, Jr., Rodriguez-Nieto S, Zhivotovsky B, Smertenko A (2005). Cysteine protease mcII-Pa executes programmed cell death during plant embryogenesis. Proc Natl Acad Sci U S A. 102: 14463-14468. [ Links ]
8. Burleigh BA, Caler EV, Webster P, Andrews NW (1997). A cytosolic serine endopeptidase from Trypanosoma cruzi is required for the generation of Ca2+ signaling in mammalian cells. J Cell Biol. 136:609-620. [ Links ]
9. Cazzulo JJ (2002). Proteinases of Trypanosoma cruzi: patential targets for the chemotherapy of Changas desease. Curr Top Med Chem. 2: 1261-1271. [ Links ]
10. Coffeen WC, Wolpert TJ (2004). Purification and characterization of serine proteases that exhibit caspase-like activity and are associated with programmed cell death in Avena sativa. Plant Cell. 16: 857-873. [ Links ]
11. Cuevas IC, Cazzulo JJ, Sanchez DO (2003). gp63 homologues in Trypanosoma cruzi: surface antigens with metalloprotease activity and a possible role in host cell infection. Infect Immun. 71:5739-5749. [ Links ]
12. Das M, Mukherjee SB, Shaha C (2001). Hydrogen peroxide induces apoptosis-like death in Leishmania donovani promastigotes. J Cell Sci. 114: 2461-2469. [ Links ]
13. de Diego JL, Katz JM, Marshall P, Gutierrez B, Manning JE, Nussenzweig V, Gonzalez J (2001). The ubiquitin-proteasome pathway plays an essential role in proteolysis during Trypanosoma cruzi remodeling. Biochemistry. 40: 1053-1062. [ Links ]
14. dos Reis FC, Judice WA, Juliano MA, Juliano L, Scharfstein J, Lima AP (2006). The substrate specificity of cruzipain 2, a cysteine protease isoform from Trypanosoma cruzi. FEMS Microbiol Lett. 259: 215-220. [ Links ]
15. El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, Ghedin E, Worthey EA, Delcher AL, Blandin G, Westenberger SJ, Caler E, Cerqueira GC, Branche C, Haas B, Anupama A, Arner E, Aslund L, Attipoe P, Bontempi E, Bringaud F, Burton P, Cadag E, Campbell DA, Carrington M, Crabtree J, Darban H, da Silveira JF, de Jong P, Edwards K, Englund PT, Fazelina G, Feldblyum T, Ferella M, Frasch AC, Gull K, Horn D, Hou L, Huang Y, Kindlund E, Klingbeil M, Kluge S, Koo H, Lacerda D, Levin MJ, Lorenzi H, Louie T, Machado CR, McCulloch R, McKenna A, Mizuno Y, Mottram JC, Nelson S, Ochaya S, Osoegawa K, Pai G, Parsons M, Pentony M, Pettersson U, Pop M, Ramirez JL, Rinta J, Robertson L, Salzberg SL, Sanchez DO, Seyler A, Sharma R, Shetty J, Simpson AJ, Sisk E, Tammi MT, Tarleton R, Teixeira S, Van Aken S, Vogt C, Ward PN, Wickstead B, Wortman J, White O, Fraser CM, Stuart KD, Andersson B (2005). The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 309: 409-415. [ Links ]
16. Ellis M, Sharma DK, Hilley JD, Coombs GH, Mottram JC (2002). Processing and trafficking of Leishmania mexicana GP63. Analysis using GP18 mutants deficient in glycosylphosphatidylinositol protein anchoring. J Biol Chem. 277: 27968-27974. [ Links ]
17. Ersfeld K, Barraclough H, Gull K (2005). Evolutionary relationships and protein domain architecture in an expanded calpain superfamily in kinetoplastid parasites. J Mol Evol. 61: 742-757. [ Links ]
18. Franke de Cazzulo BM, Martinez J, North MJ, Coombs GH, Cazzulo JJ (1994). Effects of proteinase inhibitors on the growth and differentiation of Trypanosoma cruzi. FEMS Microbiol Lett. 124: 81-86. [ Links ]
19. Freire-de-Lima CG, Nascimento DO, Soares MB, Bozza PT, Castro- Faria-Neto HC, de Mello FG, DosReis GA, Lopes MF (2000). Uptake of apoptotic cells drives the growth of a pathogenic trypanosome in macrophages. Nature. 403: 199-203. [ Links ]
20. Garcia MP, Nobrega OT, Teixeira AR, Sousa MV, Santana JM (1998). Characterisation of a Trypanosoma cruzi acidic 30 kDa cysteine protease. Mol Biochem Parasitol. 91: 263-272. [ Links ]
21. Gonzalez J, Ramalho-Pinto FJ, Frevert U, Ghiso J, Tomlinson S, Scharfstein J, Corey EJ, Nussenzweig V (1996). Proteasome activity is required for the stage-specific transformation of a protozoan parasite. J Exp Med. 184: 1909-1918. [ Links ]
22. Grutter MG (2000). Caspases: key players in programmed cell death. Curr Opin Struct Biol. 10: 649-655. [ Links ]
23. Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y (2000). A ubiquitin-like system mediates protein lipidation. Nature. 408:488-492
24. [ Links ] Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, Anupama A, Apostolou Z, Attipoe P, Bason N, Bauser C, Beck A, Beverley SM, Bianchettin G, Borzym K, Bothe G, Bruschi CV, Collins M, Cadag E, Ciarloni L, Clayton C, Coulson RM, Cronin A, Cruz AK, Davies RM, De Gaudenzi J, Dobson DE, Duesterhoeft A, Fazelina G, Fosker N, Frasch AC, Fraser A, Fuchs M, Gabel C, Goble A, Goffeau A, Harris D, Hertz- Fowler C, Hilbert H, Horn D, Huang Y, Klages S, Knights A, Kube M, Larke N, Litvin L, Lord A, Louie T, Marra M, Masuy D, Matthews K, Michaeli S, Mottram JC, Muller-Auer S, Munden H, Nelson S, Norbertczak H, Oliver K, O'Neil S, Pentony M, Pohl TM, Price C, Purnelle B, Quail MA, Rabbinowitsch E, Reinhardt R, Rieger M, Rinta J, Robben J, Robertson L, Ruiz JC, Rutter S, Saunders D, Schafer M, Schein J, Schwartz DC, Seeger K, Seyler A, Sharp S, Shin H, Sivam D, Squares R, Squares S, Tosato V, Vogt C, Volckaert G, Wambutt R, Warren T, Wedler H, Woodward J, Zhou S, Zimmermann W, Smith DF, Blackwell JM, Stuart KD, Barrell B, et al. (2005). The genome of the kinetoplastid parasite, Leishmania major. Science. 309:436-442. [ Links ]
25. Klemba M, Goldberg DE (2002). Biological roles of proteases in parasitic protozoa. Annu Rev Biochem. 71:275-305. [ Links ]
26. Kosec G, Alvarez VE, Aguero F, Sanchez D, Dolinar M, Turk B, Turk V, Cazzulo JJ (2006). Metacaspases of Trypanosoma cruzi: possible candidates for programmed cell death mediators. Mol Biochem Parasitol. 145:18-28. [ Links ]
27. Lillico S, Field MC, Blundell P, Coombs GH, Mottram JC (2003). Essential roles for GPI-anchored proteins in African trypanosomes revealed using mutants deficient in GPI8. Mol Biol Cell. 14:1182-1194. [ Links ]
28. Madeo F, Herker E, Maldener C, Wissing S, Lachelt S, Herlan M, Fehr M, Lauber K, Sigrist SJ, Wesselborg S, Frohlich KU (2002). A caspase-related protease regulates apoptosis in yeast. Mol Cell. 9:911-917. [ Links ]
29. Marino G, Uria JA, Puente XS, Quesada V, Bordallo J, Lopez- Otin C (2003). Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem. 278:3671-3678. [ Links ]
30. Martinez-Calvillo S, Lopez I, Hernandez R (1997). pRIBOTEX expression vector: a pTEX derivative for a rapid selection of Trypanosoma cruzi transfectants. Gene. 199:71-76. [ Links ]
31. Morty RE, Bulau P, Pelle R, Wilk S, Abe K (2006). Pyroglutamyl peptidase type I from Trypanosoma brucei: a new virulence factor from African trypanosomes that de-blocks regulatory peptides in the plasma of infected hosts. Biochem J. 394: 635-645. [ Links ]
32. Mottram JC, Helms MJ, Coombs GH, Sajid M (2003). Clan CD cysteine peptidases of parasitic protozoa. Trends Parasitol. 19:182-187. [ Links ]
33. Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A (2005). The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 9:769-779. [ Links ]
34. Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R (2005). A genomic and functional inventory of deubiquitinating enzymes. Cell. 123:773- 786. [ Links ]
35. Parussini F, Garcia M, Mucci J, Aguero F, Sanchez D, Hellman U, Aslund L, Cazzulo JJ (2003). Characterization of a lysosomal serine carboxypeptidase from Trypanosoma cruzi. Mol Biochem Parasitol. 131:11-23. [ Links ]
36. Piacenza L, Peluffo G, Radi R (2001). L-arginine-dependent suppression of apoptosis in Trypanosoma cruzi: contribution of the nitric oxide and polyamine pathways. Proc Natl Acad Sci U S A. 98:7301-7306. [ Links ]
37. Rawlings ND, Morton FR, Barrett AJ (2006). MEROPS: the peptidase database. Nucleic Acids Res. 34:D270-2. [ Links ]
38. Uhlmann F (2001). Secured cutting: controlling separase at the metaphase to anaphase transition. EMBO Rep. 2: 487-492. [ Links ]
39. Uren AG, O'Rourke K, Aravind LA, Pisabarro MT, Seshagiri S, Koonin EV, Dixit VM (2000). Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol Cell. 6:961-967. [ Links ]
40. Vercammen D, van de Cotte B, De Jaeger G, Eeckhout D, Casteels P, Vandepoele K, Vandenberghe I, Van Beeumen J, Inze D, Van Breusegem F (2004). Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. J Biol Chem. 279:45329-45336. [ Links ]
41. Zeledon R, Bolanos R, Rojas M (1984). Scanning electron microscopy of the final phase of the life cycle of Trypanosoma cruzi in the insect vector. Acta Trop. 41:39-43. [ Links ]
42. Zingales B, Pereira ME, Almeida KA, Umezawa ES, Nehme NS, Oliveira RP, Macedo A, Souto RP (1997). Biological parameters and molecular markers of clone CL Brener-the reference organism of the Trypanosoma cruzi genome project. Mem Inst Oswaldo Cruz. 92:811-814. [ Links ]
Received on July 27, 2006.
Accepted on August 14, 2006.














