Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
Biocell
Print version ISSN 0327-9545
Biocell vol.32 no.1 Mendoza Jan./Apr. 2008
Models for the recent evolution of protocadherin gene clusters
Marcos Morgan
Laboratorio de Genética, Departamento de Ecología, Genética y Evolución. Facultad de Ciencias Exactas y Naturales, Universidad de Buenos Aires. 4° piso, Pabellón II, Ciudad Universitaria. (1428) Buenos Aires, Argentina.
Address correspondence to: Lic. Marcos Morgan. Laboratorio de Genética, Departamento de Ecología, Genética y Evolución, Facultad de Ciencias Exactas y Naturales, Universidad de Buenos Aires, 4º piso, Pabellón II, Ciudad Universitaria. (1428) Buenos Aires, ARGENTINA. E-mail: morgan@icgeb.org
ABSTRACT: The clustered protocadherins (Pcdhs) are single-pass transmembrane proteins that constitute a subfamily within the cadherin superfamily. In mammals, they are arranged in three consecutive clusters named a, b, and g. These proteins are expressed in the nervous system and are targeted to mature synapses. Interestingly, different neurons express different subsets of isoforms; however, little is known about the functions and expression of the clustered Pcdhs.
Previous phylogenetic analyses that compared rodent and human clusters postulated the recent occurrence of gene duplication events. Using standard phylogenetic methods, I confirmed the prior observations, and I show that duplications are likely to occur through unequal crossing-over events between two, and sometimes three, different Pcdh genes. The results are consistent with the fact that these genes undergo gene conversion. Recombination events between different clustered Pcdh genes appear to underlie concerted evolution through gene conversion and gene duplications through unequal crossing-over. In this work, I provided evidence that the unit of duplication of these genes in both the mouse and the human and within each cluster is the same. The unit of duplication includes the extracellular domain-coding sequence of an isoform and its promoter along with the cytoplasmic domain-coding region of the immediately upstream isoform in the cluster.
Key words: Protocadherin; Gene birth; Gene conversion; Cluster evolution.
Introduction
Cadherins are a superfamily of cell adhesion molecules that can be divided into various families, one of which is the protocadherin (Pcdh) family. Some of the Pcdh genes are grouped in three clusters named a, b, and g (Wu and Maniatis, 1999). The human (Homo sapiens) and the mouse (Mus musculus) genomes have one of each type (Wu et al., 2001). The a and g clusters consist of a variable region, where unusually large exons (VRE) with the same orientation are arranged in tandem and followed by a constant region of three additional exons. These last exons encode an invariable Cterminal cytoplasmic region. Each VRE encodes a signal peptide, six cadherin ectodomains (ECs), a transmembrane segment, and the remaining region of the cytoplasmic domain. Every VRE has its own promoter, and the mature mRNA is synthesized via cissplicing (Tasic et al., 2002; Wang et al., 2002; Fig. 1).The b clusters are similarly organized except that they lack the three common exons. Thus, every b VRE is equivalent to a gene.

FIGURE 1. Schematic organization of a generic Pcdh cluster. VRE, variable region exon; Pcdh, protocadherin; EC, cadherin ectodomain; DCR, divergent cytoplasmic region.
The gene order of the mouse and the human a and g clusters is conserved (Wu et al., 2001; Wu, 2005). Three groups of Pcdhs can be defined within the g clusters on the basis of sequence similarity: ga, gb, and gc (Wu and Maniatis, 1999). The functional implications of the divisions are not well understood. In the mouse and the human, the gc VREs lie immediately upstream of the three common exons of the cluster and are extremely similar to the two VREs immediately upstream of the a cluster three common exons (Wu et al., 2001). Interestingly, in the ga and gb groups, proximal VREs usually have very similar sequences; however, the evolutionary relationships, which are well supported, do not seem to be the result of recent duplications.
The human b cluster contains 16 genes (PCDHB1 to PCDHB16), whereas the mouse b cluster has 22 (Pcdhb1 to Pcdhb22) (Wu et al., 2001; Vanhalst et al., 2001). The difference in the number of genes is due to numerous duplication events following the co-divergence of the lineages, combined with the loss of some genes in humans (Vanhalst et al., 2001), but the duplication mechanisms are still unknown. Phylogenetic analyses have shown that the N-terminal ectodomain of some b Pcdhs in humans are very similar (Noonan et al., 2004; Miki et al., 2005). For example, PCDHB8 and PCDHB13 have very similar third ECs. Thus, it appears that their genes emerged from a recent duplication event.
Genes in the Pcdh clusters undergo gene conversion (Noonan et al., 2004), but homogenization of the genetic information is restricted only to specific regions. Generally, most of the cytoplasmic domains and the sixth EC (EC6) sequences are very similar among the Pcdhs in each cluster. Consequently, much of the phylogenetic information is found in the N-terminal ECs. This variability in the ECs may provide specificity in cell-cell adhesion and recognition, probably through homophilic interactions.
The clustered Pcdhs are highly expressed in the nervous system and are targeted to mature synapses and intracellular compartments (Phillips et al., 2003). The complete deletion of the g Pcdh cluster in mice does not impair neurogenesis, but the cluster is required for the survival of some neuronal populations such as spinal interneurons (Wang et al., 2002). Interestingly, the different Pcdh isoforms are expressed in a punctate pattern throughout the brain (Komura et al., 1998; Frank et al., 2005). The ga and gb Pcdhs are expressed in a monoallelic and combinatorial fashion, while the expression of the gc Pcdhs is biallelic (Kaneko et al., 2006). Similarly, the a Pcdhs gene regulation is monoallelic except for the gc-like isoforms (Kaneko et al., 2006; Esumi et al., 2005).
To gain further insight into the regulation of these genes, I analyzed the recent evolution of the mouse and the human clusters. I showed that the clustered Pcdh gene duplications result from unequal crossing-over events between exons. This is consistent with the fact that the clustered genes are subject to gene conversion because both phenomena, gene duplication and gene conversion, seem to be consequences of recombination between different VREs. Moreover, I showed that the unit of duplication of these genes includes the EC-coding sequences of a VRE and its promoter along with a part of the cytoplasmic domain-coding region of the VRE immediately upstream.
Methods
All of the sequences were downloaded from GenBank (http://www.ncbi.nlm.nih.gov/) (AF152501, AF217742, AF217744-AF217749, AF217750, AF217752-AF217757, AF326296, AY013771-AY013784, AY013786-AY013791, NM_031857, NM_018900-MN_018911, AY573971-AY573983, NM_018912-NM_018921, NM_032088, NM_003735, NM_033584-NM_033595, NM_018922-NM_018927, NM_003736, NM_033574-NM_033580, NG_000012, NG_000016, and NG_000017). A pre-alignment of the whole sequences was performed to establish the limits of the regions and was followed by an alignment of the selected sequences by use of ClustalX (Thompson et al., 1997) under default parameters. I considered that each DCR (Divergent Cytoplasmic Region) coding sequence starts with the last three consecutive conserved nucleotides and ends with the VRE. For Pcdhb3 and PCDHB16, the alternative sequences were used (see below). To establish the EC limits, I considered that each domain ends after the VXVXVDXNDNAPXF-conserved motif. The unrooted trees were obtained with ClustalX (Thompson et al., 1997) by use of the neighbor-joining method under default parameters and edited with the TREEVIEW program (Page, 1996). To estimate branch supports, 1000 bootstrap replicates were performed. The nucleotide sequences were translated in silico to obtain the Pcdh amino acid sequences. Aligned sequences were shaded with the BoxShade program (http://www.ch.embnet.org/software/BOX_form.html).
The PipMaker program (http://pipmaker.bx.psu.edu/pipmaker/) (Schwartz et al., 2000) was used with the chaining option selected. Interspersed repeats were identified with the RepeatMasker program (http://www.repeatmasker.org/).
Results
Description of the clustered Pcdhs cytoplasmic domain
Because the clustered Pcdhs genes undergo gene conversion due to putative recombination events between different VREs, I examined whether unequal crossing-over between different genes could explain the emergence of the new Pcdhs in the clusters. In this case, the new Pcdhs should have N-terminal ECs similar to one isoform and a cytoplasmic domain similar to another (Fig. 2). A visual inspection of the aligned human ga Pcdhs, however, shows that although the N-terminal regions are quite variable, the C-terminal regions are extensively conserved (Fig. 3). The same is true for the other clusters (Miki et al., 2005). Thus, I limited the analysis of the C-terminal region to the last approximately 20 amino acids, which do not seem be affected by gene conversion (Fig. 4). I refer to this region as the divergent cytoplasmic region (DCR).

FIGURE 2. Models that explain the evolution of the Pcdh gene clusters. The different VRE, depicted with boxes, are identified with capital letters and divided into 5´- and 3´- regions for simplicity. The first region encodes the variable ECs and the last one the DCR. Models (A) and (C) require one unequal crossing-over event, whereas model (B) requires two. In each case, the resulting strand with the highest number of VREs is selected.

FIGURE 3. Alignment of human ga Pcdhs sequences. EC, cadherin ectodomain; DCR, divergent cytoplasmic region.

FIGURE 4. Alignment of cytoplasmic domains. (A) Alignment of human a cluster cytoplasmic domains. (B) Alignment of human b cluster cytoplasmic domains. PCDHB1 was excluded from the analysis because it has a considerably divergent sequence. DCR, divergent cytoplasmic region.
Analysis of the b cluster VREs
Since the divergence of the human and the mouse lineages, the number of b Pcdhs has increased in both species. For example, PCDHB11 and PCDHB12 ECs are very similar and are orthologs of Pcdhb19 ECs (Noonan et al., 2004; Miki et al., 2005; Additional file 1). Thus, it can be assumed that their coding sequences originated from a recent duplication event. Because their genes are contiguous in the chromosome, the duplication mechanism could have been a single unequal crossing-over event between adjacent genes, as shown in Figures 2A and 5A. According to this model, theDCR-coding sequences of PCDHB10 and PCDHB11 emerged from the same duplication event. A visual inspection of the protein sequences confirmed that these Pcdh DCRs are very similar (Fig. 6A). The same mechanism can explain the origin of PCDHB9, because it has ECs similar to those of PCDHB10 (Noonan et al., 2004; Miki et al., 2005; Additional file 1) and a DCR similar to that of the immediately downstream gene (Fig. 6A). A phylogenetic analysis of the DCR-coding regions confirms these observations (Fig. 6B).
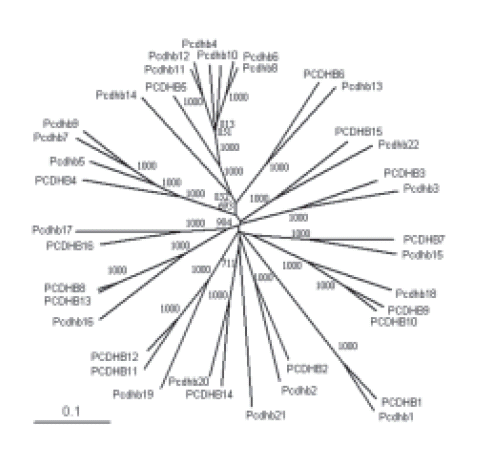
ADDITIONAL FILE 1. Evolutionary tree of the b Pcdh ECs. Unrooted evolutionary tree of the human and mouse b Pcdh ECs 1 to 4 obtained with the neighborjoining method. Bootstrap values over 500 are shown. The scale bar represents a phylogenetic distance of 0.1.

FIGURE 5. A possible scenario for the evolution of each cluster. The different VRE are represented by boxes as shown in Figure 2. The order of the events may change in some cases. (A) Evolution of part of the human b cluster. (B) Evolution of part of the murine b cluster. (C) Evolution of part of the a cluster. (D) Evolution of part of the g cluster.

FIGURE 6. Analysis of the human b cluster DCRs. (A) Alignment of human b cluster DCRs. On the left, the three pairs of proteins with very similar ECs are indicated. On the right, three pairs of proteins that have similar DCRs are indicated. The sequences follow the cluster order. Proteins encoded by upstream VREs are on top, and those encoded by downstream VREs appear at the bottom. Genetic footprints were determined manually and are boxed except for the PGKEI footprint, which is underlined. The two crosses on the PCDHB16 sequence indicate the position of two stop codons that were removed from the DNA sequence when the computational translation was performed. PCDHB1 was excluded from the analysis because it has a considerably divergent DCR. (B) Unrooted evolutionary tree of the genomic sequences encoding the human b cluster DCRs obtained using the neighbor-joining method. The PCDHB1 DCR-coding sequence was excluded from the analysis. Bootstrap values over 500 are shown. DCR-coding sequences associated by genetic footprints are indicated with parentheses. The scale bar represents a phylogenetic distance of 0.1.
As shown in Figure 6A, PCDHB7 and PCDHB12 also have similar DCRs. Because PCDHB8 and PCDHB13 have similar ECs (Noonan et al., 2004; Miki et al., 2005; Additional file 1), this observation can be explained by the mechanism proposed in Figures 2B and 5A. If the model is correct, the genomic sequence between PCDHB7 and PCDHB8 should be very similar to the sequence between PCDHB12 and PCDHB13. In fact, a comparison of both sequences with the PipMaker program (Schwartz et al., 2000) reveals a striking similarity (Fig. 7). Notably, this does not occur with the adjacent intergenic regions. The similarity between the intergenic regions downstream of PCDHB16 and PCDHB9 (Additional file 2A) and the similarity between the intergenic regions downstream of PCDHB10 and PCDHB11 (Additional file 2B) support the two first steps of the model proposed in Figure 5A.

FIGURE 7. Analysis of the human b cluster intergenic regions. Percentage identity plot obtained comparing the genomic sequence between PCDHB6 and PCDHB16 with that between PCDHB11 and PCDHB14. The horizontal axis indicates the nucleotide position of the genomic sequence between PCDHB11 and PCDHB14, and the vertical axis shows the percentage identity between the two sequences. Note that PCDHB16 is downstream of PCDHB8 and not downstream of PCDHB15. CpG/GpC>0.60, CpG island where the observed to expected CpG/GpC ratio lies between 0.6 and 0.75; CpG/GpC>0.75, CpG island where the ratio exceeds 0.75.

ADDITIONAL FILE 2. Additional analysis of the human b cluster intergenic regions. Percentage identity plots obtained comparing the intergenic regions between (A) PCDHB9 and PCDHB10, (B) PCDHB11 and PCDHB12, and (C) Pcdhb19 and Pcdhb20, with the intergenic regions between the genes indicated on the left. The horizontal axis indicates the nucleotide position of the sequences on the top, and the vertical axis shows the percentage identity between the intergenic regions between the genes indicated on the left with that on the top. CpG/GpC=0.60, CpG island where the observed to the expected CpG/GpC ratio lies between 0.6 and 0.75; CpG/GpC=0.75, CpG island where the ratio exceeds 0.75.
Next, I investigated whether the proposed mechanisms are specific to the human by analyzing the evolution of the mouse b cluster. According to the models described in Figures 2 and 5B, the ECs-coding sequence of a particular VRE is always linked to the DCR-coding sequence of the gene immediately upstream. In the mouse cluster, Pcdhb4, Pcdhb6, Pcdhb8, Pcdhb10, Pcdhb11, and Pcdhb12 have very similar ECs (Additional file 1). Thus, their coding sequences probably originate from the same ancestral gene. When the DCRs of this cluster are compared, Pcdhb3, Pcdhb5, Pcdhb7, Pcdhb9, Pcdhb10, and Pcdhb11 share a very similar footprint (Fig. 8A). This observation is consistent with the proposed models. The finding was further confirmed by a phylogenetic analysis of the DCR-coding region (Fig. 8B).

FIGURE 8. Analysis of the mouse b cluster DCRs. (A) Alignment of mouse b cluster DCRs. On the left, proteins whose ECs are orthologous to those of PCDHB5 are indicated. The sequences follow the cluster order as shown in Figure 6. The footprint common to all the DCRs whose corresponding genes are immediately upstream of genes that encode ECs orthologs of PCDHB5 ECs is boxed. The PGKEI footprint is underlined. The slash on the Pcdhb3 sequence indicates a thymine deleted from the genomic sequence to introduce a reading frame shift when the computational translation was performed. Pcdhb1 was excluded from the analysis because it has a considerably divergent DCR. (B) Unrooted evolutionary tree of the genomic sequences that encode the mouse b cluster DCRs obtained using the neighbor-joining method. The Pcdhb1 DCR-coding sequence was excluded from the analysis. Bootstrap values over 500 are shown. DCR-coding sequences immediately upstream of genes that encode ECs orthologs of PCDHB4 and PCDHB5 ECs are indicated with parentheses. The scale bar represents a phylogenetic distance of 0.1.
To gain further insight into the evolution of this cluster, I analyzed the intergenic regions. A comparative phylogenetic analysis of the human and the mouse ectodomains reveals that the PCDHB5 ECs are orthologs of those in Pcdhb4, Pcdhb6, Pcdhb8, Pcdhb10, Pcdhb11, and Pcdhb12 and that PCDHB4 ECs are orthologs to those of Pcdhb5, Pcdhb7, and Pcdhb9 (Vanhalst et al., 2001; Additional file 1). According to the model proposed in Figure 5B, the intergenic region separating PCDHB4 and PCDHB5 is orthologous to the intergenic regions between Pcdhb3 and Pcdhb4, Pcdhb5 and Pcdhb6, Pcdhb7 and Pcdhb8, Pcdhb9 and Pcdhb10, Pcdhb10 and Pcdhb11, and Pcdhb11 and Pcdhb12. Also, the intergenic region separating PCDHB3 and PCDHB4 is orthologous to the intergenic regions between Pcdhb4 and Pcdhb5, Pcdhb6 and Pcdhb7, and Pcdhb8 and Pcdhb9. When the sequences are compared using the PipMaker program (Schwartz et al., 2000), only those that appear to be orthologous show a greater than 50% similarity (Fig. 9). This supports the idea that the unit of duplication for these genes consists of the DCR-coding region of a Pcdh gene along with the promoter and EC-coding region of the gene immediately downstream. The similarity between the intergenic regions downstream of Pcdhb10 and Pcdhb11 (Additional file 3C) supports the last step in the model shown in Figure 5B. The similarity between the intergenic regions downstream of Pcdhb5 and Pcdhb7 (Additional file 3A), together with the similarity between the intergenic regions downstream of Pcdhb6 and Pcdhb8 (Additional file 3B), supports the fourth step of the model. The first three steps of the model could not be validated by analyzing the non-coding sequences due to the lack of phylogenetic signal. Thus, other possible mechanisms cannot be formally excluded. Nevertheless, gene duplication through unequal crossing-over does not seem to be specific to a particular species.

FIGURE 9. Analysis of the mouse b cluster intergenic regions. Comparison of the human intergenic regions between PCDHB3 and PCDHB5 with the mouse intergenic regions between Pcdhb2 and Pcdhb13. The horizontal axis represents the length of the human intergenic regions. The vertical axis represents the similarity between the human intergenic region (above) and the mouse intergenic region between the genes indicated on the left. Supposedly paralogous sequences are indicated. CpG/GpC≥ 0.60, CpG island where the observed to expected CpG/GpC ratio lies between 0.6 and 0.75; CpG/GpC≥ 0.75, CpG island where the ratio exceeds 0.75.

ADDITIONAL FILE 3. Additional analysis of the mouse b cluster intergenic regions. Percentage identity plots obtained comparing the intergenic regions between (A) Pcdhb5 and Pcdhb6, (B) Pcdhb6 and Pcdhb7, and (C) Pcdhb10 and Pcdhb11, with the intergenic regions between the genes indicated on the left. The horizontal axis indicates the nucleotide position of the sequence on the top, and the vertical axis shows the percentage identity between the intergenic regions indicated on the left with that on the top. CpG/GpC=0.60, CpG island where the observed to the expected CpG/ GpC ratio lies between 0.6 and 0.75; CpG/GpC=0.75, CpG island where the ratio exceeds 0.75.
Analysis of the a cluster VREs
Subsequently, I investigated whether these mechanisms are common to the different clusters. Thus, I conducted a similar analysis with the a Pcdhs. Although duplications in these clusters did not occur after the separation of the rodent and the human lineages, it has been suggested that PCDHA6 and PCDHA7, and PCDHA8 and PCDHA9 are duplications of the same gene pair, because PCDHA6 is very similar to PCDHA8, and PCDHA7 to PCDHA9 (Wu et al., 2001). I proposed an alternative model to explain the observations, namely, a recombination event between two VREs, as shown in Figures 2C and 5C. This model is supported by the short DCR shared by PCDHA5 and PCDHA7 (Fig. 10A). The association was validated by a phylogenetic analysis of the DCR-coding sequence (Fig. 10B). Similar results were obtained analyzing the rat a cluster (Additional file 4). Also, the similarity between the introns downstream of PCDHA5 and PCDHA7 (Fig. 11A) and between the introns downstream of PCDHA6 and PCDHA8 (Fig. 11B) support the model. Consequently, the mechanisms do not appear to be species-or cluster-specific.

FIGURE 10. Analysis of the human a cluster DCRs. (A) Alignment of human a cluster DCRs. On the left, the two pairs of proteins whose genes are associated by a putative duplication event are indicated. On the right, one pair of proteins that share the same genetic footprint is indicated. The sequences follow the cluster order as shown in Figure 6. The gc-like Pcdhs were excluded from the analysis. The genetic footprint was determined manually and is boxed. (B) Unrooted evolutionary tree of the genomic sequences that encode the human a cluster DCRs obtained with the neighbor-joining method. DNA sequences of the gc-like Pcdhs were excluded from the analysis. Bootstrap values over 500 are shown. DCR-coding sequences associated by the same genetic footprint are indicated with a parenthesis. The scale bar represents a phylogenetic distance of 0.1.

ADDITIONAL FILE 4. Analysis of the rat a cluster DCRs. (A) Alignment of rat a cluster DCRs. On the left, the two pairs of proteins whose genes are associated by a putative duplication event are indicated. On the right, pairs of proteins that share genetic footprints are indicated. The sequences follow the cluster order as shown in Figure 6. The gc-like Pcdhs were excluded from the analysis, and the first constant amino acids were included. Genetic footprints were determined manually and are boxed. (B) Unrooted evolutionary tree of the genomic sequences that encode the rat a cluster DCRs obtained using the neighbor-joining method. DNA sequences of the gc-like Pcdhs were excluded from the analysis. Bootstrap values over 500 are shown. DCR-coding sequences associated by genetic footprints are indicated with parentheses. The scale bar represents a phylogenetic distance of 0.1.

FIGURE 11. Analysis of the human a cluster introns. Percentage identity plots obtained by comparing the introns between (A) PCDHA5 and PCDHA6, and (B) PCDHA6 and PCDHA7 with the introns between the exons indicated on the left. The horizontal axis indicates the nucleotide position of the intron on the top, and the vertical axis shows the percentage identity between the introns indicated on the left with that on the top. CpG/GpC≥0.60, CpG island where the observed to the expected CpG/GpC ratio lies between 0.6 and 0.75; CpG/GpC≥0.75, CpG island where the ratio exceeds 0.75.
Analysis of the g cluster VREs
First, I analyzed the ga group VREs. There are 12 ga Pcdhs (Pcdhgas) in humans (PCDHGA1 to PCDHGA12) and in mice (Pcdhga1 to Pcdhga12). PCDHGA2 and PCDHGA3 as well as PCDHGA8 and PCDHGA9 are very similar (Wu and Maniatis, 1999; Wu et al., 2001). In this group, the third and fourth ECs have the highest paralogous diversity at synonymous sites and are the least affected by gene conversion (Noonan et al., 2004). Thus, I tested if the evolutionary relationships between the above-mentioned Pcdhs were retained in these ECs. The results of the neighbor-joining method indicate that PCDHGA2 and PCDHGA3 are grouped in the evolutionary tree, and the bootstrap support of the corresponding branch is strong (approximately 93%). PCDHGA8 and PCDHGA9 are also grouped with the highest bootstrap value for the resulting branch. Consequently, the evolutionary relationships are significant. All other associations are considerably weaker (Additional file 5A). For example, the strongest of them among PCDHGA8, PCDHGA9, and PCDHGA10 has a bootstrap support of approximately 73%. A previous report showed that PCDHGAn and Pcdhgan are very similar (V n ∟ N; n ≤ 12) (Wu et al., 2001). Thus, essentially the same results were obtained when the analysis was repeated in mice (Additional file 5B).
To determine if the models presented in Figure 2 can explain the evolution of this group of Pcdhs, I analyzed the DCR of these genes. Again, the expected results were obtained. PCDHGA1 and PCDHGA2 have similar DCRs, and the same occurs with PCDHGA7 and PCDHGA8 (Fig. 12). Also, similar results were obtained for the mouse cluster (Additional file 6).

ADDITIONAL FILE 5. Evolutionary trees of the human and the mouse ga group ECs. Unrooted evolutionary tree of the (A) human and (B) mouse ga group ECs 3 and 4, obtained with the neighbor-joining method. Bootstrap values over 500 are shown. The scale bar represents a phylogenetic distance of 0.1.

FIGURE 12. Analysis of the human ga group DCRs. (A) Alignment of the human ga group DCRs. On the left, pairs of proteins with very similar third and fourth ECs are indicated. On the right, pairs of proteins that share genetic footprints are indicated. The sequences follow the cluster order as shown in Figure 6. Genetic footprints were determined manually and are boxed. (B) Unrooted evolutionary tree of the genomic sequences that encode the human ga group DCRs obtained using the neighbor-joining method. Bootstrap values over 500 are shown. DCR-coding sequences associated by genetic footprints are indicated with parentheses. The scale bar represents a phylogenetic distance of 0.1.
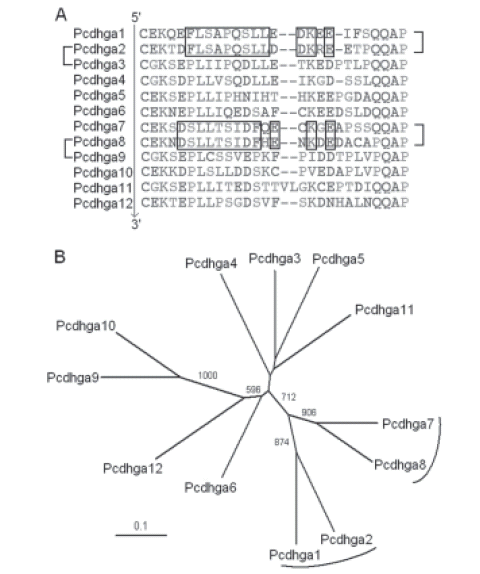
ADDITIONAL FILE 6. Analysis of the mouse ga group DCRs. (A) Alignment of the mouse ga group DCRs. On the left, pairs of proteins with very similar third and fourth ECs are indicated. On the right, pairs of proteins that share genetic footprints are indicated. The sequences follow the cluster order as shown in Figure 6. The first constant amino acids were included. Genetic footprints were determined manually and are boxed. (B) Unrooted evolutionary tree of the genomic sequences that encode the mouse ga group DCRs obtained with the neighbor-joining method. Bootstrap values over 500 are shown. DCR-coding sequences associated by genetic footprints are indicated with parentheses. The scale bar represents a phylogenetic distance of 0.1.
An analysis of the gb group ECs showed that PCDHGB4 and PCDHGB5 have very similar ectodomains and that the same is true for the PCDHGB6 and PCDHGB7 ECs (Additional file 7A). The PCDHGB6 cytoplasmic domain, however, is more similar to those of PCDHGB4 and PCDHGB5 than to that of PCDHGB7 (Additional files 8A and 9). The same is true for the murine gb group (Additional files 7B and 8B). These observations can be explained by the model shown in Figure 5D. The intronic phylogenetic signal of the cluster, although consistent with the proposed models, is low (Additional file 10). Therefore, the simple model cannot be further validated, and other mechanisms for the evolution of the cluster cannot be formally excluded.

ADDITIONAL FILE 7. Evolutionary trees of the human and the mouse gb group ECs. Unrooted evolutionary tree of (A) the human and (B) the mouse gb group ECs 2, 3, and 4, obtained with the neighbor-joining method. Bootstrap values over 500 are shown. The scale bar represents a phylogenetic distance of 0.1.

ADDITIONAL FILE 8. Evolutionary trees of the human and the mouse gb group DCRs. Unrooted evolutionary tree of (A) the human and (B) the mouse gb group DCR-coding sequences obtained with the neighbor-joining method. Bootstrap values over 500 are shown. The scale bar represents a phylogenetic distance of 0.1.
ADDITIONAL FILE 9. Alignment of the human gb Pcdhs sequences. (A) Alignment of the human gb Pcdhs sequences. (B) Alignment of the gb Pcdh C-terminal amino acids. Residues shared by PCDHGB4, PCDHGB5, and PCDHGB6 are indicated in bold.

ADDITIONAL FILE 10. Analysis of the human g cluster introns. Percentage identity plots obtained comparing the introns between (A) PCDHGA8 and PCDHGB5, (B) PCDHGB5 and PCDHGA9, and (C) PCDHGA2 and PCDHGA3, with the introns between the exons indicated on the left. The horizontal axis indicates the nucleotide position of the intron on the top, and the vertical axis shows the percentage identity between the introns indicated on the left with that on the top. CpG/GpC=0.60, CpG island where the observed to the expected CpG/GpC ratio lies between 0.6 and 0.75; CpG/GpC=0.75, CpG island where the ratio exceeds 0.75.
Discussion
The Pcdh clusters, together with other clusters in the mammalian genomes, undergo concerted evolution. Two clear indicators of this process are the reduced synonymous diversity among paralogs and the increase in the CG content of the third codon (Galtier et al., 2001). Usually, almost the entire length of each Pcdh cytoplasmic domain is subject to gene conversion (Noonan et al., 2004) so that divergence among paralogs is reduced to narrow regions (Figs. 3 and 4). Something similar appears to happen in the a cluster first EC (EC1) sequences in humans, where just a few non-conserved amino acids are surrounded by more than 30 conserved amino acids to each side (see Miki et al., 2005). This does not seem to occur by chance, because the homologous amino acids in mice are conserved and overlap an RGD motif located in a loop homologous to the quasi-b helix conformation of N-cadherin (Shapiro et al., 1995; Morishita et al., 2006). Nevertheless, the divergent sequences can still provide important information about the function and evolution of the Pcdhs.
The various clusters are subject to different evolutionary pressures. Two of the clusters, a and b, appear to have undergone duplications quite recently, and it is not possible to clearly resolve their complete phylogeny due to a loss of signal with time. Consequently, I was only able to analyze the genes that emerged recently. In the human b cluster, the DCR coding sequences were clearly duplicated together along with their immediately downstream Pcdh gene. The g clusters seem to be under the influence of specific evolutionary forces that physically constrained them because there is no evidence of recent duplication events in the analyzed species. This indicates that these clusters operate as supra-organizations above the gene level.
Recombination between Pcdh genes is likely to be a frequent event because these genes are subject to gene conversion (Noonan et al., 2004; Miki et al., 2005). Therefore, recombination between different genes probably underlies both gene duplication and concerted evolution. On the basis of the evidence presented and the mechanisms proposed, I suggest possible scenarios for the evolution of the different clusters in Figure 5. In this study, I provided evidence that shows that the unit of evolution of the Pcdh genes consists of the EC-coding region and the promoter of a certain VRE along with the DCR-coding sequence of the VRE immediately upstream.
This fact appears to be the rule and seems to apply even to the particular duplication that generated PCDHB8 and PCDHB13. Considering that PCDHB14 and Pcdhb20 ECs are orthologs (Additional file 1), the fact that PCDHB13 and Pcdhb19 share the PGKEI footprint in their DCRs is consistent with the proposed model (see Figs. 6A, 8A, and Additional file 2C). Note that, because most of the phylogenetic information is in the 5'-region of the VREs, the orthology of the new recombinant isoforms was usually based on just the most informative part of the sequences.
Classic cadherins are cell adhesion molecules that participate in different signaling pathways. For instance, N-cadherin is a single-pass transmembrane protein that is expressed in the nervous system and regulates dendritic arborization in hippocampal neurons through its interaction with b-catenin (Junghans et al., 2005). Moreover N-cadherin can be cleaved by metalloproteinases and g-secretase, resulting in the liberation of its cytoplasmic domain as a signaling molecule (Junghans et al., 2005).
The g Pcdhs are targeted to both sides of mature synapses, presumably to modulate the strength of the synaptic union (Phillips et al., 2003). Recently, it was shown that the g cluster cytoplasmic domains can be released when the Pcdhs are inserted into the plasma membrane via g-secretase-dependent processing, after which the domains translocate to the nucleus (Hass et al., 2005; Hambsch et al., 2005). Moreover, the cytoplasmic constant region of the g cluster can activate the transcription of reporter genes downstream of different g Pcdh promoters (Hambsch et al., 2005); however, all the g Pcdhs are not expressed at the same time (Kohmura et al., 1998; Frank et al., 2005). In this study, I showed that the different Pcdh cleavage products can have different phylogenetic origins. Notably according to the proposed models, the cytoplasmic domain that enters the nucleus evolved together with the immediately downstream promoter region and not with its own promoter. This leaves some interesting questions and suggests some hypotheses about the role of the DCR. To begin with, does the variability in the cytoplasmic region play some role? Also, does the DCR provide some kind of specificity to the induction of the Pcdh locus? For example, it is possible that the cytoplasmic domain preferentially activates the immediately downstream gene.
Conclusions
Comparative analysis between genomic sequences of different species can provide information about the regulation of a particular gene or cluster. Understanding the recent evolution of a particular genomic region may provide further insight into the mechanisms of regulation. In this study, I have provided evidence that duplication events in the Pcdh clusters occur through unequal crossing-over between two and sometimes three different VREs. Interestingly, the unit of duplication always consisted of the EC-coding sequence and the promoter of a certain isoform along with the DCR-coding region of the isoform immediately upstream in the cluster. The reorganization of the clusters after the duplications events suggests new hypotheses about the regulation of the clusters that must be tested experimentally.
Acknowledgements
I thank Drs. P. Cramer and V.A. Confalonieri for discussions and critical comments on the manuscript.
References
1. Esumi S, Kakazu N, Taguchi Y, Hirayama T, Sasaki A, Hirabayashi T, Koide T, Kitsukawa T, Hamada S, Yagi T (2005). Monoallelic yet combinatorial expression of variable exons of the protocadherin-a gene cluster in single neurons. Nat Genet. 37: 171-176. [ Links ]
2. Frank M, Ebert M, Shan W, Phillips GR, Arndt K, Colman DR, Kemler R (2005). Differential expression of individual gamma-protocadherins during mouse brain development. Mol Cell Neurosci. 29: 603-616. [ Links ]
3. Galtier N, Piganeau G, Mouchiroud D, Duret L (2001). GC-content evolution in mammalian genomes: the biased gene conversion hypothesis. Genetics. 159: 907-911. [ Links ]
4. Haas IG, Frank M, Véron N, Kemler R (2005). Presenilin-dependent processing and nuclear function of g-protocadherins. J Biol Chem. 280: 9313-9319. [ Links ]
5. Hambsch B, Grinevich V, Seeburg PH, Schwarz MK (2005). g- protocadherins, presenilin-mediated release of c-terminal fragment promotes locus expression. J Biol Chem. 280: 15888-15897. [ Links ]
6. Junghans D, Haas IG, Kemler R (2005). Mammalian cadherins and protocadherins: about cell death, synapses and processing. Curr Opin Cell Biol. 17: 446-452. [ Links ]
7. Kaneko R, Kato H, Kawamura Y, Esumi S, Hirayama T, Hirabayashi T, Yagi T (2006). Allelic gene regulation of protocadherin-a and -g clusters involving both monoallelic and biallelic expression in single Purkinje cells. J Biol Chem. 281: 30551-30560. [ Links ]
8. Kohmura N, Senzaki K, Hamada S, Kai N, Yasuda R, Watanabe M, Ishii H, Yasuda M, Mishina M, Yagi T (1998). Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron. 20: 1137-1151. [ Links ]
9. Miki R, Hattori K, Taguchi Y, Tada MN, Isosaka T, Hidaka Y, Hirabayashi T, Hashimoto R, Fukuzako H, Yagi T (2005). Identification and characterization of coding single-nucleotide polymorphisms within human protocadherin-a and -b gene clusters. Gene. 349: 1-14. [ Links ]
10. Morishita H, Umitsu M, Murata Y, Shibata N, Udaka K, Higuchi Y, Akutsu H, Yamaguchi T, Yagi T, Ikegami T (2006). Structure of the CNR/protocadherina first cadherin domain reveals diversity across cadherin families. J Biol Chem. 281: 33650-33663. [ Links ]
11. Noonan JP, Grimwood J, Schmutz J, Dickson M, Myers RM (2004). Gene conversion and the evolution of protocadherin gene cluster diversity. Genome Res. 14: 354-366. [ Links ]
12. Page RDM (1996). TREEVIEW: An application to display phylogenetic trees on personal computers. Comput Appl Biosci. 12: 357-358. [ Links ]
13. Phillips GR, Tanaka H, Frank M, Elste A, Fidler L, Benson DL, Colman DR (2003). g-protocadherins are targeted to subsets of synapses and intracellular organelles in neurons. J Neurosci. 23: 5096-5104. [ Links ]
14. Schwartz S, Zhang Z, Frazer KA, Smit A, Reiner C, Bouck J, Gibbs R, Hardison R, Miller W (2000). PipMaker-A web server for aligning two genomic DNA sequences. Genome Res. 10: 577-586. [ Links ]
15. Shapiro L, Fannon AM, Kwong PD, Thompson A, Lehmann MS, Grübel G, Legrand J-F, Als-Nielsen J, Colman DR, Hendrickson WA (1995). Structural basis of cell-cell adhesion by cadherins. Nature. 374: 327-337. [ Links ]
16. Tasic B, Nabholz CE, Baldwin KK, Kim Y, Rueckert EH, Ribich SA, Cramer P, Wu Q, Axel R, Maniatis T (2002). Promoter choice determines splice site selection in protocadherin a and g pre-mRNA splicing. Mol Cell. 10: 21-33. [ Links ]
17. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876-4882. [ Links ]
18. Vanhalst K, Kools P, Eynde EV, van Roy F (2001). The human and murine protocadherin-b one-exon gene families show high evolutionary conservation, despite the difference in gene number. FEBS. 495: 120-125. [ Links ]
19. Wang X, Su H, Bradley A (2002). Molecular mechanisms governing Pcdh-g gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes & Dev. 16: 1890-1905. [ Links ]
20. Wang X, Weiner JA, Levi S, Craig AM, Bradley A, Sanes JR (2002). Gamma protocadherins are required for survival of spinal interneurons. Neuron. 36: 843-854. [ Links ]
21. Wu Q, Maniatis T (1999). A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 97: 779-790. [ Links ]
22. Wu Q, Zhang T, Cheng J-F, Kim Y, Grimwood J, Schmutz J, Dickson M, Noonan JP, Zhang MQ, Myers RM, Maniatis T (2001). Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res. 11: 389-404. [ Links ]
23. Wu Q (2005). Comparative genomics and diversifying selection of the clustered vertebrate protocadherin genes. Genetics. 169: 2179-2188. [ Links ]
Received on October 24, 2006.
Accepted on August 22, 2007.














