Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Anales de la Asociación Química Argentina
Print version ISSN 0365-0375
An. Asoc. Quím. Argent. vol.93 no.4-6 Buenos Aires Jan./Dec. 2005
REGULAR PAPERS
Qsar Modeling Of Anti-Hiv-1 Activity Of Hept Derivatives. Optimization Of Correlation Weights Of Morgan Extended Connectivity In Graph Of Atomic Orbitals
Castro, E.A.1; Toropov, A.A.2; Toropova, A.P.2; Mukhamedzhanove, D.V.2
1INIFTA, Theoretical Chemistry Division, Chemistry Department, Faculty of Exact Sciences, La Plata National University, Suc.4, C.C. 16, La Plata, B1906ZAA, Argentina. Fax: +54-221-425-4642, Email: jubert@arnet.com.ar
2 Uzbekistan Academy of Sciences, Geology & Geophysics Institute, 700041, Khodzhibaev, 49 Street,Tashkent, Uzbekistan
Received August 12th, 2005. In final form August 29th, 2005
Dedicated to the memory of the late Prof. Hans J. Schumacher on the occasion of his 100th birthday
Abstract
Quantitative Structure - Activity Relationships (QSAR) for Anti-HIV-1 activity of 1-[2- hydroxyethoxy-methyl]-6-(phenylthio) thymine] (HEPT) derivatives have been obtained by optimization of correlation weights of local invariants in Labelled Hydrogen - Filled Graphs (LHFGs) and Graph of Atomic Orbitals (GAOs). The best model is based on correlation weights of Morgan extended connectivity of first order in the GAO. Statistical characteristics of this model are the following: n = 59, R2 = 0.91, s = 0.41, F = 577 (Training Set); n = 20, R2 = 0.91, s = 0.42, F = 183 (Test Set).
Resumen
Se han determinado las Relaciones Cuantitativas Estructura-Actividad (QSAR) para la actividad Anti-HIV-1 de los derivados del 1-[2-hidrometoxi-metil]-6-(feniltio)timina (HEPT) por medio de la optimización de los pesos de correlación de los invariantes locales de los Grafos Completos de Hidrógeno Señalados (LHFGs) y de los Grafos de los Orbitales Atómicos (GAOs). El mejor de los modelos obtenidos basado en los pesos de correlación de la conectividad extendida de Morgan del primer orden es el GAO. Las características estadísticas de este modelo son las siguientes: n = 59, R2 = 0,91, s = 0,41, F = 577 (Conjunto de Entrenamiento); n = 20, R2 = 0,91 ,s = 0,42, F = 183 (Conjunto de Prueba).
Introduction
The Acquired Immuno Deficiency Syndrome (AIDS) epidemic has taken more than 3 million lives in 2004, and an estimated 5 million people have acquired the Human Immunodeficiency Virus (HIV) in 2004, bringing to 42 million people the number of people globally living with the virus. HIV/AIDS has become the most devastating pandemic in the recorded contemporary history. It has killed 40 million people in the last 20 years and the World Health Organization has estimated that there would be up to 100 million new infections in the next 10 years [1]. Because human type immunodeficiency virus (HIV-1) is the causative agent of AIDS, extensive research clinical works are currently going on to try to block its replication [2]. Quantitative structure- property/activity relationships (QSPR/QSAR) may be used as a tool for prediction anti-HIV-1 activity. From a literature survey it was found that faced with the need to develop effective anti- HIV drugs, QSAR studies have been used on potential anti-HIV drug candidates acting on different targets [3-13].
There are number of ways to represent molecular structures in order to employ the QSPR/ QSAR analyses, such as the hydrogen - suppressed graph (i.e., hydrogen atoms are ignored), hydrogen-filled graph, 3D molecular model, and quantum chemical models. In the present study two versions of elucidation of molecular structure have been used. These are the Labelled Hydrogen-Filled Graph (LHFG) and Graph of Atomic Orbitals (GAO). GAO is an attempt to take into account the structure of atoms in QSPR/QSAR analyses. Recently, we have obtained satisfactory predictions of physical chemistry properties and biological activities within the realm of QSPR/QSAR theory via molecular graphs representation by means of optimization of correlation weights of local graph invariants (OCWLI) [14-23].
The present study is aimed to estimate the capability of the OCWLI to predict numerical values of the Anti-HIV-1 activity, in log (1/LC50) taken from Ref. [2] (see Fig. 1).
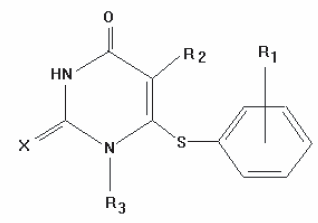
Figure 1. General structure of HEPT derivatives studied.
Method
The descriptors used in the present study are defined as
| (1) |
where G is the LHFG or GAO, Vk stands for a chemical element in case of the LHFG or Atomic Orbital in case of the GAO. The xECk is the Morgan extended connectivity of x-th order calculated with the recursion formula [14-23]
| (2) |
The correlation coefficient between the 0XCW(G,xEC) and log(1/LC50) is a function of the correlation weights (CWs). Under such circumstances the following steps can be taken [14-23]:
1. Using the Monte Carlo method, the values of the Cws giving rise to the largest possible values of the correlation coefficient can be calculated;
2. Using the least squares method, the following equation may be obtained with compounds of the training set
| (3) |
3. Predictive capability of Eq.(3) may be validated for compounds belonging to a test set.
The conversion of the LHFG into the corresponding GAO may be carried out by the following scheme:
i) Each vertex of the LHFG is replaced by a group of Atomic Orbitals (AOs). Such groups of AOs on all atoms under consideration are listed in Table 1.
Table 1. AO groups on atoms under consideration.

ii) The element aij of adjacency matrix of the GAO is defined as 1 if the i-th and j-th vertices of the GAO fall in groups of different atoms from the LHFG and these atoms have joint edge in the LHFG; otherwise, the aij = 0.
The LHFG and GAO on compounds No 49 (training set) are demonstrated in Fig 2 and Fig.3, respectively.
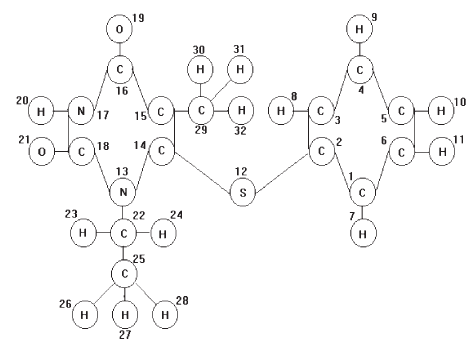
Figure 2. The LHFG of compound No 49 (training set).

Figure 3. The GAO for Compound No 49 (training set)
Results and Discussion
Table 2 lists statistical characteristics of the OCWLI models of Anti-HIV-1 activity. One can see from the Table 2 that best OCWLI model of the activity under consideration takes place for the 0X(GAO,1EC). Table 3 lists results of three probes OCWLI based on the 0X(GAO,1EC). Numerical values of the CWs are displayed in Table 4. Calculation of the 0X(GAO,1EC) for the 49th compound from the training set is illustrated in Table 5. The results of this calculation are based on CWs obtained for the first OCWLI probe from the Table 4.
Table 2. Number of OCWLI parameters and statistical characteristics of QSAR on different versions of flexible descriptor calculated by Eq.(1).

Table 3. Statistical characteristics of three OCWLI probes to model of Anti-HIV-1 activity based on 0X(GAO,1EC)

Table 4. Correlation weights for calculating of the 0X(GAO,1EC) obtained in three OCWLI probe s

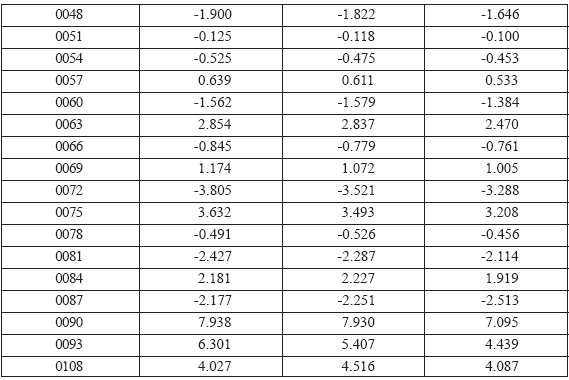
Table 5. Calculation of 0XCW(GAO,1EC) for compound No 49 (training set) with correlation weights of first OCWLI probe. The 0XCW(GAO,1EC) is equal to 17.285


Observed values of the Anti-HIV-1 activity from Ref. [2], numerical values of the 0X(GAO,1EC), and structure compounds of general formula shown in Fig.1 are listed in Table 6. OCWLI model of log(1/LC50) calculated as
| (4) |
are also shown in Table 6. Statistical features of Eq.(4) can be seen from the Table 3 (first OCWLI probe).
Table 6. Chemical Structures of the Compounds Studied and Their Anti-HIV-1 Activity (log (1/EC50))(see Fig. 1)

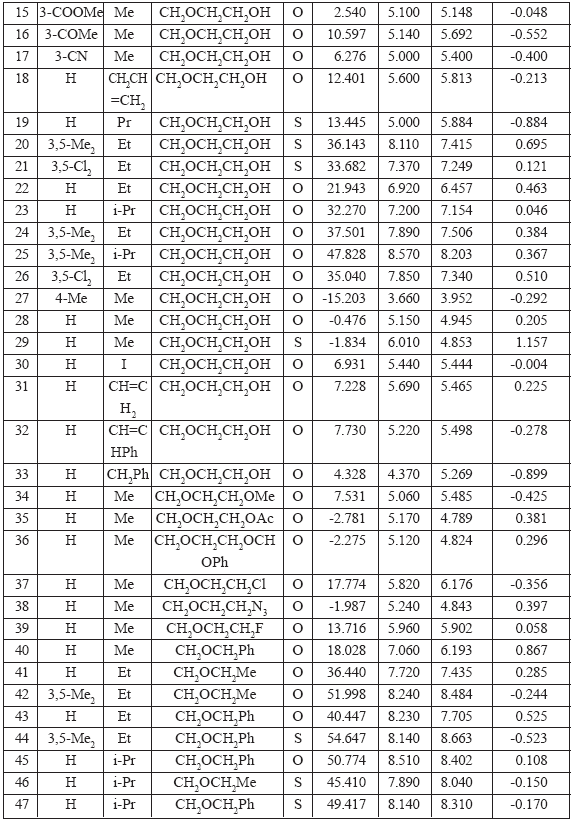

According to Ref. [2], the 79 HEPT derivatives under consideration have been studied by means of neural networks technique and via a multiple linear regression. Standard errors in this previous research examined in Ref. [2] fall within an interval going from 0.607 till 0.185. Thus statistics data associated with Eq.(4), i.e., 0.41 - 0.42 may be considered as reasonably satisfactory.
Conclusions
The optimization of correlation weights of local invariants in graph of atomic orbitals may be used as a useful tool for predicting numerical values of Anti-HIV-1 activity. This sort of molecular descriptor belongs to the category of flexible molecular descriptors and they have shown to be quite suitable for QSAR/QSPR studies to predict several kinds of physicochemical properties and biological activities [24-29]. The advantages and usefulness of this method can be summarized as follows:
- The method is relatively simple to be implemented in QSAR/QSPR studies.
- Numerical accuracy is satisfactory and it gives a solid basis for interpretative purposes in order to assess the main factors governing biological activities and physicochemical properties.
- The comparison between rigid and flexible molecular descriptors shows that the last category of molecular descriptor yields better predictive results.
- Present results on Anti-HIV-1 activity makes a significative contribution with regard to several previous studies on this issue.
- Main conclusions derived from this research allow us to extend present analysis in order to study other molecules having potential Anti-HIV-1 activity. At present some complementary calculations are under development and results will be presented elsewhere in the forthcoming future.
References
[1] Mwau, M.; McMichel, A.J., J. Gene Med., 2003, 5, 3-10. [ Links ]
[2] Douali, L.; Villemin, D.; Cherqaoui, D., J. Chem. Inf. Comput. Sci., 2003, 43, 1200- 1207. [ Links ]
[3] Castro, E.A.; Torrens, F.; Toropov, A.A.; Nesterov, I.V.; Nabiev, O.M., Molec. Simul., 2004, 30 (10), 691-696. [ Links ]
[4] Garg, R.; Gupta, S.P.; Gao, H.; Babu, M.S.; Debnath, A.K.; Hansch, C., Chem. Rev., 1999, 99, 3525-3601. [ Links ]
[5] H. Bazoui, M. Zahouily, S. Boulajaaj, S. Sebti, D. Zakarya, SAR-QSAR Environ. Res., 13 (2002), 567-577. [ Links ]
[6] Luco, J.M.; Ferretti, F.H., J. Chem. Inf. Comput. Sci., 1997, 37, 392-401. [ Links ]
[7] Katritzky, A.R.; Oliferenko, A.; Lomaka, A.; Karelson, M., Bioorg. Med. Chem. Lett., 2002, 12, 3453-3457. [ Links ]
[8] Gayathri, P.; Pande, V.; Sivakumar, R.; Gupta, S.P., Bioorg. Med. Chem., 2001, 9,3059-3063. [ Links ]
[9] Pungpo, P.; Hannongbua, S., J. Mol. Graph. Model., 2000, 18,581-590. [ Links ]
[10] Siddiqui, A.Q.; McGuigan, C.; Ballatore, C.; Zuccotto, F.; Gilbert, I.H.; De Clercq, E.; Balzarini, J., J. Med. Chem., 1999, 42, 4122-4128. [ Links ]
[11] Huang, X.; Xu, L.; Luio, X., Fan, K.; Ji, R.; Pei, G.; Chen, K.; Jiang, H., J. Med. Chem., 2002, 45, 333-343. [ Links ]
[12] Chan, J.H.; Hong, J.S.; Hunter III, R.N.; Orr, G.F.; Cowan, J.L.; Sherman, D.L.; Sparks, S.M.; Reitter, B.E.; Andrews III, C.W.; Hazen, R.J.; St. Clair, M.; Boone, L.R.; Ferris, R.G., Creech, K.L.; Roberts, G.B.; Short, S.A.; Weaver, K.; Ott, R.J.; Ren, J.; Hopkins, A., Stuart, D.I., Stammers, D.K., J. Med. Chem., 2001, 44,1866- 1882. [ Links ]
[13] Leonard, J.T.; Roy, K., QSAR Comb. Sci., 2004, 23, 23-35. [ Links ]
[14] Toropov, A.A.; Toropova, A.P., J. Mol. Struct. (THEOCHEM), 2001, 538, 197-199. [ Links ]
[15] Toropov, A.A.; Toropova, A.P., J. Mol. Struct. (THEOCHEM), 2001, 538, 287-293. [ Links ]
[16] Toropov, A.A.; Toropova, A.P., J. Mol. Struct. (THEOCHEM), 2002, 581, 11-15. [ Links ]
[17] Toropov, A.A.; Toropova, A.P., J. Mol. Struct. (THEOCHEM), 2003, 637, 1-10 [ Links ]
[18] Toropov, A.A.; Toropova, A.P., Internet Electron. J. Mol. Des., 2002, 1, 109-113, http://www.biochempress.com . [ Links ]
[19] Peruzzo, P.J., Marino, D.J.G.; Castro, E.A.; Toropov, A.A., Internet Electron. J. Mol. Des., 2003, 2, 334-347, http://www.biochempress.com . [ Links ]
[20] Toropov, A.A.; Toropova, A.P.; Nesterov, I.V.; Nabiev, O.M., J. Mol. Struct., (THEOCHEM) 2003, 640, 175-181. [ Links ]
[21] Toropov, A.A.; Toropova, A.P.; Nesterova, A.I.; Nabiev, O.M., Chem. Phys. Letters, 2004, 384, 357-363. [ Links ]
[22] Toropov, A.A., Roy, K., J. Chem. Inf. Comput. Sci., 2004, 44,179-186. [ Links ]
[23] Toropov, A.A.; Benfenati, E., J. Mol. Struct. (THEOCHEM), 2004, 676, 165-169. [ Links ]
[24] Toropov, A.A.; Duchowicz, P.R., Castro, E.A., Molecules, 2004, 9, 1019-1033. [ Links ]
[25] Castro, E.A.; Toropova, A.P., Toropov, A.A., Mekhamedjanova, D.V., Struct. Chem., 2005, 16(3), 311-330. [ Links ]
[26] Castro, E.A., Toropov, A.A., Nesterova, A.I., Nabiev, O.M., Cent. Eur. J. Chem., 2004, 2(3), 1-24. [ Links ]
[27] Duchowicz, P.R.; Castro, E.A., Toropov, A.A., Nesterova, I.V.; Nabiev, O.M. Molec. Diversity 2004, 8(4), 325-330. [ Links ]
[28] Duchowicz, P.R., Castro, E.A.; Toropov, A.A.; Nestrova, A.I.; Nabiev, O.M., J. Argent. Chem. Soc., 2004, 92(1-3), 29-42. [ Links ]
[29] Castro, E.A.; Duchowicz, P.R., Toropov, A.A.; Nestrova, A.I.; Nazarov, A.U., J. Theor. Comput. Chem., 2004, 3(1), 31-41. [ Links ]














