Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
Revista de la Sociedad Entomológica Argentina
Print version ISSN 0373-5680On-line version ISSN 1851-7471
Rev. Soc. Entomol. Argent. vol.68 no.1-2 Mendoza Jan./June 2009
Genetic analysis of a recently detected urban population of Lutzomyia evansi (Diptera: Psychodidae)in Colombia
Análisis genético de una población urbana de Lutzomyia evansi (Diptera: Psychodidae), recientemente detectada en Colombia
Bejarano, Eduar Elías*, Winston Rojas**, Sandra Uribe***, Iván Darío Vélez**** and Charles H. Porter*****
*Grupo de Investigaciones Biomédicas, Universidad de Sucre, Carrera 14 No. 16 B-32, Apartado Aéreo 406, Sincelejo, Colombia; e-mail: eduarelias@yahoo.com
**Grupo de Genética Molecular, Universidad de Antioquia, Calle 62 No. 52-69, Apartado Aéreo 1226, Medellín, Colombia.
***Grupo de Sistemática Molecular, Universidad Nacional de Colombia, Calle 59A No. 63-20, Medellín, Colombia.
****Programa de Estudio y Control de Enfermedades Tropicales, Universidad de Antioquía, Calle 62 No. 52-59, Apartado Aéreo 1226, Medellín, Colombia.
*****Division of Parasitic Diseases, National Center for Infectious Diseases, Centers for Disease Control and Prevention, 1600 Clifton Rd, Atlanta, GA 30333, USA.
ABSTRACT. Lutzomyia evansi (Núñez-Tovar) is the vector of the parasite Leishmania infantum in rural zones of Northern Colombia. An attempt was made to determine the origin of a recently detected urban population of Lutzomyia evansi by genetically characterizing specimens from seven geographically distinct localities in the Colombian Caribbean. Insect specimens were collected in rural and urban environments of areas endemic for visceral leishmaniasis or free of the disease. Nine polymorphic sites, nine nucleotide haplotypes and a single aminoacid haplotype were found within the 315 bp fragment sequenced, corresponding to the 3' end of the cytochrome b mitochondrial gene. Paired genetic distances between the haplotypes, estimated with the Kimura two-parameters model, varied from 0,0032-0,0194. Analysis revealed low genetic variability between specimens from urban and rural localities. Several of the sand flies collected in the city of Sincelejo (department of Sucre), where autochthonous visceral leishmaniasis cases have appeared in recent years, were genetically similar to those of a rural focus of the disease (El Contento, on the neighboring department of Córdoba). The epidemiological implications of this finding for Leishmania infantum transmission in the Colombian Caribbean are discussed.
KEY WORDS. Sand flies; Genetics; Lutzomyia evansi; Visceral leishmaniasis; Colombia.
RESUMEN. Lutzomyia evansi (Núñez-Tovar) es el insecto transmisor del parásito Leishmania infantum en zonas rurales del norte de Colombia. Con el propósito de establecer el probable origen de una población urbana del vector, detectada en años recientes, se caracterizaron genéticamente ejemplares de Lutzomyia evansi de siete localidades geográficas del Caribe Colombiano. Los flebotomíneos fueron recolectados en ambientes rurales y urbanos de zonas endémicas y no endémicas de leishmaniasis visceral. Dentro del fragmento secuenciado de 315 pb correspondiente al extremo 3' del gen mitocondrial citocromo b, se encontraron nueve sitios polimórficos, nueve haplotipos nucleotídicos y un solo haplotipo aminoacídico. Las distancias genéticas pareadas entre los haplotipos, estimadas con el modelo de Kimura de dos parámetros, oscilaron entre 0,0032 y 0,0194. El análisis reveló la existencia de una baja variabilidad genética entre especímenes de localidades urbanas y rurales. Varios de los flebotomíneos recolectados en la zona urbana de la ciudad de Sincelejo, departamento de Sucre, donde en años recientes aparecieron casos autóctonos de leishmaniasis visceral, fueron genéticamente similares a los de El Contento, en el cercano departamento de Córdoba, foco rural de la enfermedad. Se discuten las implicaciones epidemiológicas de este hallazgo para la transmisión de Leishmania infantum en el Caribe Colombiano.
PALABRAS CLAVE. Flebotomíneos; Genética; Lutzomyia evansi; Leishmaniasis visceral; Colombia.
Recibido: 28-11-2008;
Aceptado: 15-05-2009
INTRODUCTION
Visceral leishmaniasis (VL), a re-emergen zoonosis in the Americas (Arias et al., 1996 Feliciangeli et al., 1999; Enserink, 2000), i caused by the trypanosomatid Leishmani infantum Nicolle. VL is the most sever of three clinical forms of leishmaniasis generally producing death in childre under five if left untreated. Formerly, VL wa present only in rural areas but now occur in urban and periurban habitats of Brazi Venezuela and Colombia (Arias et al., 1996 Aguilar et al., 1998; Bejarano et al., 2002 Cortés & Fernández, 2008). This may b attributed to both human migration an climatic or environmental changes that affec populations of vectors, some of which hav adapted to anthropogenic environment (Walsh et al., 1993).
In South and Central America Lutzomyi longipalpis (Lutz & Neiva), L. cruz (Mangabeira) and L. evansi (Núñez-Tovar are the known vectors of Le. infantum (Trav et al., 1990; Ferro et al., 1995; Santos et al 1998). The last of these transmits the parasit in Colombia and Venezuela (Travi et al 1990; Feliciangeli et al., 1999). In 1999 L. evansi was detected in an urban area o the Colombian Caribbean (Bejarano et al 2001), the first cases of VL appearing two years later in the city of Sincelejo (Bejarano et al., 2002). An entomological survey of dwellings in a neighborhood where the affected children lived revealed the presence of L. evansi (Bejarano et al., 2002). Lutzomyia longipalpis has not been found, to date, in this neighborhood despite intensive entomological sampling.
This investigation was undertaken to determine the probable geographical origin of this recently detected urban population of the vector. Thus, specimens of L. evansi from rural and urban localities in Colombia were characterized using the mitochondrial cytochrome b gene. Mitochondria have matrilineal inheritance, which permits the geographical dispersion of females to be traced. Females have greater epidemiological importance than males through being directly involved in transmission of the parasite (Bejarano, 2001).
MATERIALS AND METHODS
Collection and identification of sand flies. The study was carried out in seven localities of the Caribbean coastal region of Colombia, each of which presents a distinct epidemiological profile for VL (Table I). Sand flies were collected in and around houses, using CDC light traps and Shannon traps respectively. The head, one wing and the genitalia of each specimen were removed to determine the species using the taxonomic key of Young & Duncan (1994), and Galati (2003).
DNA Extraction. DNA was extracted using the protocol of Collins et al. (1987) with the following modifications. Each specimen was macerated in 60 µl of lysis buffer (0,08 NaCl, 0,16 M saccharose, 0,06 M EDTA, 0,5% SDS, 0,1 M Tris-HCL at pH 7,5) and incubated at 65°C for 30 min, after which 14 µl of 8 M potassium acetate was added and the mixture incubated on ice for 30 min. It was then centrifuged at 12 000 x g for 10 min and the supernatant transferred to a new vial. The DNA was precipitated for 18 h with 200 µl of absolute ethanol at -20°C and centrifuged at 12 000 x g for 20 min. After decanting off the supernatant, the DNA was rinsed with 200 µl of 70% ethanol and then with 200 µl of absolute ethanol. Finally, it was left to dry at room temperature and re-suspended in 30 µl of ultrapure water.
DNA amplification. A segment of the mitochondrial DNA genome was amplified by PCR using the forward primer [5'-CA(T/C)ATTCAACC(A/T)GAATGATA-3'] and the reverse primer [5'-GGTA(C/T)(A/T) TTGCCTCGA(T/A)TTCG(T/A)TATGA-3'] (Ready et al., 1997). This region includes the 3' end of the cytochrome b gene. PCR amplifications were performed in a 50 µl reaction mix containing 1x PCR buffer (Promega Corporation, Madison, WI), 1,5mM MgCl2, 0,2mM of deoxynucleoside triphosphate mix (Promega), 0,3|uM of each primer, 1,5 U of Taq DNA polymerase (Promega) and 6 \úof DNA template. The cycling conditions included an initial 3-min denaturation step at 94°C, followed by 35 cycles of 1-min denaturation at 93°C, 1-min primer annealing at 50°C, and 1-min elongation at 72°C. Final extension was done at 72°C for 10 min, followed by a hold step at 4°C.
Nucleotide sequencing. PCR products separated by 1,5% agarose gel electrophoresis were visualized by ethidium bromide staining, purified by using Wizard® PCR Preps DNA Purification System Kit (Promega), and sequenced in both directions using the Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA). Sequences were determined on an ABI Prism® 377 automated DNA sequencer (Applied Biosystems) and assembled using the SeqMan program (DNASTAR, Inc., Madison, WI). The consensus nucleotide sequences were derived from 14 individuals of L. evansi. GenBank accession numbers for the sequence data are EF457569-EF457582 (Table I).
Table I. Collection sites of Lutzomyia evansi in the Caribbean coast of Colombia and Genbank accession numbers of the DNA sequences obtained in this study.

Genetic analysis. The gene that codes for the protein cytochrome b was selected for genetic analysis, since this presents the highest rate of substitution between the sequenced mitochondrial segments of Lutzomyia spp. (Bejarano, 2001). The segment analyzed is homologous with the region between positions 11 271 and 11 585 of the Aedes albopictus (Skuse) mitochondrial genome (Genbank accession no. AY072044). Nucleotide sequences were aligned with the Clustal W program running within the software package DAMBE 4.2.13 (Xia & Xie, 2001). Pairwise sequence alignments and protein translation were performed with MEGA version 3.1 (Kumar et al., 2003). To determine the phylogenetic relationships of L. evansi populations, a neighbor-joining tree was constructed on the basis of the evolutionary distances calculated by the Kimura two-parameter model (Kimura, 1980) using MEGA software (Kumar et al., 2003).
RESULTS
A total of nine nucleotide haplotypes of the cytochrome b gene was obtained from L. evansi specimens collected on the Caribbean coast of Colombia. These are the first nucleotide sequences reported for this sand fly vector species. The mean nucleotide composition of the cytochrome b gene was 9,9% guanine, 16,2% cytosine, 33,6% adenine and 40,3% thymine. Nine polymorphic and 306 conserved sites were found on this 315-bp segment of this gene (Table II). Most substitutions were transitions, five involving cytosine-thymine and three adenine-guanine. Only one corresponded to an adenine-thymine transversion. All the changes were silent, reflected in the presence of a single haplotype aminoacid among sand flies from the seven geographical locations analyzed.
Table II. Distribution of haplotypes of Lutzomyia evansi derived from the 3' end of the cytochrome b gene across Colombian localities. SI, Sincelejo; EC, El Contento; IF, Isla Fuerte; LV, Los Vidales; LP, Loma de Piedra; EA, Escobar Arriba; SR, Santa Rosa de Lima.

Three of the nine nucleotide haplotypes observed appeared in the individuals of L. evansi from the city of Sincelejo. The number of nucleotide differences between haplotypes varied from 1-6 (0,32 and 1,94%), with an overall mean of 2,78 substitutions (0,89%). The first haplotype was shared between recently urbanized sand flies (Sincelejo) and specimens from a rural focus of VL at El Contento in the neighboring department of Cordoba. The second haplotype was observed in urban insects (Sincelejo) and the sand flies from Isla Fuerte, a small island 11 km off the Caribbean coast of Colombia. The third haplotype included individuals of all three localities (Sincelejo, Isla Fuerte and El Contento). The remaining six haplotypes corresponded exclusively to sand flies collected in rural areas.
The paired genetic distances between each of the nine haplotypes, estimated using the Kimura two-parameter model, had low values of 0,0032-0,0194 (Table III). The highest value was recorded among haplotypes of rural locations that were VL foci, while the lowest ones appeared when haplotypes of rural and urban villages were paired. Two clusters were observed in the neighbor-joining tree (Fig. 1). The first cluster grouped haplotypes of urban, rural and island locations, while the second included those of three dissimilar rural villages from areas endemic or not for VL.
Table III. Kimura's two-parameter genetic distance among nine nucleotide haplotypes of Lutzomyia evansi of Colombia. All haplotype numbers correspond to Table II.

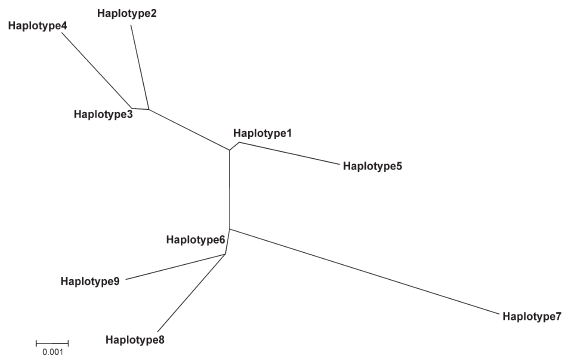
Fig. 1. Phylogenetic analysis of haplotypes of Lutzomyia evansi from Colombian localities based on a 315-bp fragment of the cytochrome b gene. Distances and groupings were determined by the Kimura's two-parameter and neighbor-joining method with the MEGA software. All haplotype numbers correspond to Table II.
DISCUSSION
The present study represents the first analysis of intraspecific genetic variability of Lutzomyia evansi in Colombia, where previous genetic studies of sand flies have been focused on L. longipalpis, L. shannoni (Dyar), and other species of the L. verrucarum group (Cárdenas et al., 2001; Uribe et al. 2001; Testa et al., 2002; Pérez-Doria et al., 2008). Analysis of the 3' end of this gene revealed low genetic variability among L. evansi isolates from the Colombian Caribbean. Genetic distance values were very low, similar to those observed with the same molecular marker in populations of L. longipalpis and L. whitmani (Antunes & Coutinho) (Ready et al., 1997; Hodgkinson et al., 2003). However, unlike these latter species, in which each nucleotide haplotype was present in a single population, several L. evansi haplotypes were shared by rural and urban individuals. Furthermore, on the phylogenetic tree the mitochondrial haplotypes of urban sand flies did not appear on distinct clusters from those of rural insects, suggesting genetic homogeneity between the two groups.
The urban sand fly population of Sincelejo proved to be genetically related to the rural one of El Contento, an established VL focus since the 1980s (Vélez et al., 1988). Several of the sand flies of both geographical localities shared the same nucleotide haplotype, clearly showing a common genetic origin. This allows us to suggest that the urban population of L. evansi could have originated by dispersion from this rural focus of the disease, taking into account that these localities are approximately 13km apart. There are no apparent geographical barriers that would impede such dispersion. However, the previous existence of the vector in urban areas could also be possible, with the current population being descended from sand flies that survived the selection pressure of antimalaria spraying campaigns carried out in the region during the 1980s.
It is difficult to determine whether vector dispersion contributed to that of the parasite, since the latter could also have arrived in infected mammal reservoirs. In recent years families displaced by politically motivated violence have arrived in Sincelejo from rural foci endemic for VL, bringing dogs with them that might be infected with Le. infantum and completing the elements required to establish a new microfocus of infection (Pérez-Doria et al., 2006). Sporadic autochthonous cases of VL have appeared since 1999 (Bejarano et al., 2002), when L. evansi was first detected in Sincelejo (Bejarano et al. 2001). This species was absent from all previous insect collections made in the city, which supports the hypothesis that it was a recent invader. Thus Lutzomyia evansi could be increasing the original limits of the Le. infantum transmission focus on the Caribbean coast of Colombia, which would explain the presence of new VL cases in areas recently colonized by the vector (Bejarano et al., 2002; Cortés & Fernández, 2008). This should be taken into account by public health entities when designing VL prevention and control measures in this country.
ACKNOWLEDGEMENTS
This work received financial support from Colciencias (grant no. 115-05-106-97) and the Universidad de Antioquia (grant no. CPT0019). The authors are grateful to Suljey Cochero for providing specimens from Loma de Piedra and Escobar Arriba, department of Sucre and to the reviewers for suggestions improving this paper.
LITERATURE CITED
1. AGUILAR, C. M., E. FERNÁNDEZ, R. FERNÁNDEZ, D. C. CANNOVA, E. FERRER, Z. CABRERA, W. J. S. SOUZA & S. G. COUTINHO. 1998. Urban visceral leishmaniasis in Venezuela. Memórias do Instituto Oswaldo Cruz 93 (1): 15-16. [ Links ]
2. ARIAS, J. R., P. S. MONTEIRO & F. ZICKER. 1996. The reemergence of visceral leishmaniasis in Brazil. Emerging Infectious Disease 2 (2): 145-146. [ Links ]
3. BEJARANO, E. E. 2001. Nuevas herramientas para la clasificación taxonómica de los insectos vectores de leishmaniosis: utilidad de los genes mitocondriales. Biomédica 21 (2): 182-191. [ Links ]
4. BEJARANO, E. E., S. URIBE, W. ROJAS & I. D. VÉLEZ. 2001. Presence of Lutzomyia evansi, a vector of American visceral leishmaniasis, in an urban area of the Colombian Caribbean coast. Transactions of the Royal Society of Tropical Medicine and Hygiene 95 (1): 27-28. [ Links ]
5. BEJARANO, E. E., S. URIBE, W. ROJAS & I. D. VÉLEZ. 2002. Phlebotomine sand flies (Diptera: Psychodidae) associated with the appearance of urban leishmaniasis in the city of Sincelejo, Colombia. Memórias do Instituto Oswaldo Cruz 97 (5): 645-647. [ Links ]
6. CÁRDENAS, E., L. E. MUNSTERMANN, O. MARTÍNEZ, D. CORREDOR & C. FERRO. 2001. Genetic variability among populations of Lutzomyia (Psathyromyia) shannoni (Dyar 1929) (Diptera: Psychodidae: Phlebotominae) in Colombia. Memórias do Instituto Oswaldo Cruz 96 (2): 189-196. [ Links ]
7. COLLINS, F. H., M. A. MÉNDEZ, M. O. RASMUSSEN, P. C. MEHAFFEY, N. J. BESANSKY & V. FINNERTY. 1987. A ribosomal RNA gene probe differentiates member species of the Anopheles gambiae complex. The American Journal of Tropical Medicine and Hygiene 37 (1): 37-41. [ Links ]
8. CORTÉS, L. A. & J. J. FERNÁNDEZ. 2008. Especies de Lutzomyia en un foco urbano de leishmaniasis visceral y cutánea en El Carmen de Bolívar, Bolívar, Colombia. Biomédica 28 (3): 433-440. [ Links ]
9. ENSERINK, M. 2000. Has leishmaniasis become endemic in the U.S.? Science 290 (5498): 1881-1883. [ Links ]
10. FELICIANGELI, M. D., N. RODRÍGUEZ, Z. DE GUGLIELMO & A. RODRÍGUEZ. 1999. The re-emergence of American visceral leishmaniasis in an old focus in Venezuela. II. Vectors and parasites. Parasite 6 (2): 113-120. [ Links ]
11. FELSENSTEIN, J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39 (4): 783-791. [ Links ]
12. FERRO, C., A. C. MORRISON, M. TORRES, R. PARDO, M. L. WILSON & R. B. TESH. 1995. Age structure, blood-feeding behavior, and Leishmania chagasi infection in Lutzomyia longipalpis (Diptera: Psychodidae) at an endemic focus of visceral leishmaniasis in Colombia. Journal of Medical Entomology 32 (5): 618-629. [ Links ]
13. GALATI, E. A. B. 2003. Morfologia, terminologia de adultos e identificação dos táxons da América. En: Rangel, E. F. & R. Lainson R (eds.), Flebotomíneos do Brasil, Editora Fiocruz, Rio do Janeiro, pp. 53-175. [ Links ]
14. HODGKINSON, V. H., J. BIRUNGI, M. QUINTANA, R. DIETZE & L. E. MUNSTERMANN. 2003. Mitochondrial cytochrome b variation in populations of the visceral leishmaniasis vector Lutzomyia longipalpis across eastern Brazil. The American Journal of Tropical Medicine and Hygiene 69 (4): 386-392. [ Links ]
15. KIMURA, M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16 (2): 111-120. [ Links ]
16. KUMAR, S., K. TAMURA & M. NEI. 2004. MEGA 3.1: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics 5 (2): 150-163. [ Links ]
17. PÉREZ-DORIA, A. J., E. E. BEJARANO & P. J. BLANCO-TUIRÁN. 2006. Presencia de Lutzomyia dubitans (Sherlock, 1962) (Diptera: Psychodidae: Phlebotominae) en la Ciudad de Sincelejo, Departamento de Sucre, Colombia. Boletín de la Asociación Española de Entomología 30 (1-2): 197-200. [ Links ]
18. PÉREZ-DORIA, A., E. E. BEJARANO, D. SIERRA & I. D. VÉLEZ. 2008. Molecular evidence confirms the taxonomic separation of Lutzomyia tihuiliensis from Lutzomyia pia (Diptera: Psychodidae) and the usefulness of pleural pigmentation patterns in species identification. Journal of Medical Entomology 45 (4): 653-659. [ Links ]
19. READY, P. D., J. C. DAY, A. A. DE SOUZA, E. F. RANGEL & C. R. DAVIES. 1997. Mitochondrial DNA characterization of populations of Lutzomyia whitmani (Diptera: Psychodidae) incriminated in the peri-domestic and silvatic transmission of Leishmania species. Bulletin of Entomological Research 87: 187-195. [ Links ]
20. SANTOS, S. O., J. ARIAS, A. A. RIBEIRO, M. P. HOFFMAN, R. A. FREITAS & M. A. F. MALACCO. 1998. Incrimination of Lutzomyia cruzi as a vector of American visceral leishmaniasis. Medical and Veterinary Entomology 12 (3): 315-317. [ Links ]
21. TESTA, J. M., J. MONTOYA-LERMA, H. CADENA, M. OVIEDO & P. D. READY. 2002. Molecular identification of vectors of Leishmania in Colombia: Mitochondrial introgression in the Lutzomyia townsendi series. Acta Tropica 84 (3): 205-218. [ Links ]
22. TRAVI, B. L., I. D. VÉLEZ, L. BRUTUS, I. SEGURA, C. JARAMILLO & J. MONTOYA. 1990. Lutzomyia evansi, an alternate vector of Leishmania chagasi in a Colombian focus of visceral leishmaniasis. Transactions of the Royal Society of Tropical Medicine and Hygiene 84 (5): 676-677. [ Links ]
23. URIBE, S., T. LEHMANN, E. D. ROWTON, I. D. VÉLEZ & C. PORTER. 2001. Speciation and population structure in the morphospecies Lutzomyia longipalpis (Lutz & Neiva) as derived from the mitochondrial ND4 gene. Molecular Phylogenetics and Evolution 18 (1): 456-461. [ Links ]
24. VÉLEZ, I. D., G. GHYSAIS, J. MARULANDA, D. A. MAYA, I. RIVERA, M. A. GUERRERO, M. I. HURTADO, J. M. CADAVID, A. OCAMPO, B. ARBELAEZ & M. WOLFF. 1988. Leishmaniasis tegumentaria americana: encuesta epidemiológica en una comunidad indígena. Iatreia 1 (1): 29-33. [ Links ]
25. WALSH, J. F., D. H. MOLYNEUX & M. H. BIRLEY. 1993. Deforestation: effects on vector-borne disease. Parasitology 106 (Suppl. 1): S55-S75. [ Links ]
26. XIA, X. & Z. XIE. 2001. DAMBE. Software package for data analysis in molecular biology and evolution. Journal of Heredity 92 (4): 371-373. [ Links ]
27. YOUNG, D. G. & M. A. DUNCAN. 1994. Guide to the identification and geographic distribution of Lutzomyia sand flies in Mexico, the West Indies, Central and South America (Diptera: Psychodidae). Memories of the American Entomological Institute 54: 1-881. [ Links ]














