Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista veterinaria
versión On-line ISSN 1669-6840
Rev. vet. vol.30 no.1 Corrientes jun. 2019
http://dx.doi.org/10.30972/vet.3013906
TRABAJO DE INVESTIGACIÓN
Molecular genetic diversity and relationship of indigenous sheep breeds of Pakistan based on nuclear microsatellite loci
Hussain, T.1; Musthafa, M.M.2; Babar, M.E.1; Shaheen, M.3; Marikar, F.M.4
1Dep. Molec. Biol.Univ. Pakistan, Lahore, Pakistan.
2Inst.Biol.Sci.Univ.Malaya, Kuala Lumpur, Malaysia.
3Univ.Vet.Anim.Sci. Lahore. 4Gen.Sir.J.Kotel.Def.Univ.Sri Lanka. E-mail: faiz.marikar@fulbrightmail.org
DOI: http://dx.doi.org/10.30972/vet.3013906
Abstract
Sheep genetic resources are high in Pakistan with a number of different breeds spread throughout the country. However, the molecular diversity of sheep breeds is poorly understood in Pakistan. Therefore, in the present study 16 microsatellite markers were used in microsatellite on Buchi and Hashtnagri indigenous breeds of Punjab and Balochistan provinces, respectively. Blood samples from 25 unrelated individuals were collected for genetic diversity and relationship investigation. The mean number of alleles on Buchi and Hashtnagri were 3.375±1.455 and 3.50±1.591, respectively. The mean observed heterozygosity for Buchi was 0.878±0.204 while for Hashtnagri it was 0.885±0.218. The mean Shannon Index showed 1.032±0.371 and 1.070±0.412 for Buchi and Hashtnagri respectively. Inbreeding estimates (FIS and FIT) showed negative values while mean gene flow showed 10.09 and mean population difference (FST) showed 2.4%. According to these results, Buchi and Hashtnagri indigenous breeds showed considerable amount of genetic diversity. There is a decent scope for conservation, effective improvement, and designing suitable breeding strategies for sheep breeds in near future.
Key words: sheep, Buchi, Hashtnagri, genetic diversity, microsatellite markers, Pakistan.
Resumen
En Pakistán los recursos genéticos ovinos son abundantes, disponiéndose de varias diferentes castas extendidas en todo el país. No obstante, la diversidad molecular de las razas de ovejas es poco conocida en la región. Por ello, en el presente estudio se investigaron 16 marcadores microsatelitales de las razas indígenas Buchi y Hashtnagri, en las provincias de Pendjab y Balochistan respectivamente. Para indagar la diversidad genética se obtuvieron muestras de sangre de 25 animales de cada raza. El promedio del número de alelos en Buchi y Hashtnagri fue de 3,375±1,455 y 3,50±1,591 respectivamente. Los promedios de heterocigosis registradas fueron de 0,878±0,204 para la raza Buchi y 0,885±0,218 para Hashtnagri. Los índices de Shannon fueron 1,032±0,371 para Buchi y 1,070±0,412 para Hashtnagri, respectivamente. La estimación de los coeficientes de endogamia (FIS y FIT) mostró valores negativos, en tanto que el flujo genético (migración) fue de 10,09 y el promedio de la diferencia poblacional (FST) resultó de 2,4%. En ambas razas, el índice de información polimórfica fue 0,56, indicando el valor del marcador del tablero. Entre las ovejas Buchi y Hashtnagri, la distancia genética estándar de Nei (Ds) fue 0,0218. Según estos resultados, ambas razas mostraron considerable diversidad genética. Los datos obtenidos permiten vislumbrar una promisoria mejoría en el área de la conservación y en el diseño de las estrategias para la cría de ovejas en el futuro cercano.
Palabras clave: oveja, Buchi, Hashtnagri, diversidad genética, marcadores microsatelitales, Pakistán.
Recibido: 23 de noviembre de 2017
Aceptado: 9 de marzo de 2018
INTRODUCTION
Livestock sector of Pakistan has some staggering numbers where it covers more than half of the agricultural gross domestic products and it stands at fifth largest milk producer in the world 1, 4. Moreover, the country is the tenth largest sheep producer in the world; endowed with nearly 34 breeds of sheep (Ovis aries) and not only forming the backbone of the rural economy of poor farmers but also add immense richness to the genetic diversity to the already rich gene pool 5.
Characterization and evaluation of genetic polymorphism among these breeds is necessary for the effective utilization of animal genetic resource rich Pakistan through suitable breeding and conservation strategies. Nevertheless, very limited efforts have been made to document their genetic diversity and conservation strategies which may lead to lose of some very valuable resources 1.
Genetic diversity is defined as variability of alleles in a population 6. Genetic characterization is the primary step in any conservation of genetic resources 7, 11. In general, the genetic variation is the core for evolution and giving the ability cope with changing environments 12. Moreover, proper assessment of genetic diversity both within and among populations is key to formulate management and conservation strategies 13.
The animal genetic diversity studies have been accelerated considerably over the last two decades with the application of molecular techniques to evolutionary studies 14. Genetic characterization is carried out in livestock researches using various molecular biology techniques like restriction fragment length polymorphism, protein polymorphism, randomly amplified polymorphic DNA, amplified fragment length polymorphismi, Mitochondrial DNA, short tandem repeat (microsatellites) and single nucleotide polymorphism 15, 20.
Microsatellite markers have been widely used as a choice of marker over the other markers to analyze the genetic variation in cattle, sheep and goats 21, 26 due to the similarities between their chromosomes 27, 28. Microsatellite markers are widely used for measuring genetic distance among closely related populations from various countries 29.
These microsatellites have become widely applied for several types of studies, due to their advantages over other markers; the relative ease in obtaining markers, neutrality, high polymorphisms, easy automation of analytical procedure 30. It detects variation in simple nucleotide repeats, random and abundant distribution across the genome and co-dominant as well 31, 35. Therefore, the current study was designed to genetically characterize the selected two sheep breeds of Pakistan using microsatellite markers. Buchi and Hashtnagri breeds of sheep were selected for characterization and this is the first effort of this nature on these indigenous but valuable breeds. Buchi is inhabitant of Cholistan desert i.e. Bahawalpur, Bahawalnagar, Rahim YarKhan, and hot regions of southern
Punjab including Multan and Muzaffargarh districts of the Punjab province, while Hashtnagri is located in Mardan, Peshawar, Haripur, Bannu and Kohat districts of Khyber Pakhtunkhwa, Province of Pakistan 34, 36. Hashtnagri is categorized as fat-tail sheep breed of Pakistan and they possess short legs with a small switch hanging over the fat-tail. Generally Hashtnagri has a medium-sized and white coated head with a partially or wholly black or tan in body colour. They have some excellent abilities in producing carpet quality wool with a fiber diameter of 35 μm 36. Buchi breeds is described as compact, medium sized, white coat with black spots on body but totally black ones also can be found. It’s a thin tailed type of sheep breed mainly reared for wool and mutton 34.
The main objective of the study was to characterize and observe genetic variation in sheep breeds through microsatellite markers for making affective strategies for the conservation and proper management of the biological resources of Pakistan.
MATERIAL AND METHODS
Animal selection and DNA isolation. Blood samples (10 ml) were collected from Buchi and Hashtnagri sheep into 200 μl EDTA containing tubes aseptically from jugular vein puncture. Unrelated Buchi sheep samples (n=25) were collected from Government Livestock Experiment Station, Jugaitpeer, District Bhawalpur, Punjab province and surrounding areas while Hashtnagri sheep samples (n=25) were taken from District Peshawar and surrounding areas in Khyber Pakhtunkhwa province. Genomic DNA was extracted using inorganic method of extraction as used by 37. The final concentration of extracted DNA was brought to 50 ng/μL through spectrophotometry and 0.8% agarose gel electrophoresis.
Markers selection and genotyping. The International Society of Animal Genetics described more than 1400 microsatellite markers that have been identified in cattle and out of these, more than 500 markers can be amplified efficiently in sheep 28. From this list, sixteen (n=16) microsatellite markers were selected for the amplification in Buchi and Hashtnagri breeds of Pakistan (Table 1). All microsatellites were optimized for PCR amplification through BioRad thermo cycler using reaction mixture of 25 μL contained 100 ng DNA, 50 mM KCl, 10 mM Tris-HCl, 2 mM dNTPs, 1.5 mM MgCl2, 1.25 pmol/ μL of forward and reverse primers and 0.15 μL of 5U Taq polymerase (Frementas, USA). Touch-down PCR was used with the initial denaturing at 95°C for 4 minutes, followed by 35 cycles each for 30 seconds at 94°C for denaturation, 45 seconds at 64°-54° C for annealing and 45 seconds at 72°C for extension. Final extension was given from 10 minutes at 72°C. The products were run on 12% non denaturing polyacrylamide gel in 1X TAE buffer at 120 volts for seven hours. Statistical analysis. Polyacrylamide gel electrophoresis results were analyzed by the relative flow method. Statistical analysis for calculation of genetic variability measures such as number of alleles, expected and observed heterozygosity, homozygosity and Polymorphic Information Content (PIC) was calculated for all microsatellite markers. Genetic distances between the breeds were calculated according to Nei, 1973 38.
Table 1. Microsatellite markers used in this study with allele size range and chromosome number.
The unweighted pair group method was used to make the dendrogram of genetic distance. Popgene 1.31 39 and Power Stat software were used for calculations. The usual F-statistics parameters were measured for these two breeds using Popgene software. FIS is known as inbreeding co-efficient which is defined as inbreeding co-efficient of an individual (I) relative to the subpopulation (S) and FIT is measured as a co-efficient of an individual (I) in relation to the population.
FIS displays value between -1 and +1, where positive values indicate heterozygote deficiency (inbreeding) and negative values depicts heterozygosity excess (out breeding) 40. FST is a measure of the effect of population subdivision, which is the reduction in subpopulation heterozygosity due to genetic drift 41.
Unlike the other F-estimates, FST always shows positive values, ranging between 0 and 1. FST is widely used to calculate gene flow in population genetic studies: 0.25*(1-FST)/FST. This highly versatile parameter is even used as a genetic distance measure between two populations instead of a fixation index among many populations 42.
RESULTS
A total of 54 and 56 alleles were found in the 16 loci tested on Buchi and Hashtnagri breed respectively (Table 2).
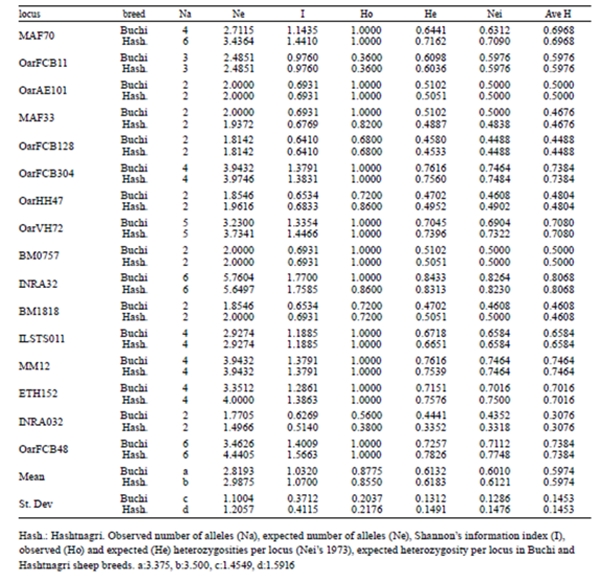
Table 2. Summary of genetic variation statistics for 16 loci used in Buchi and Hashtnagri sheep breeds.
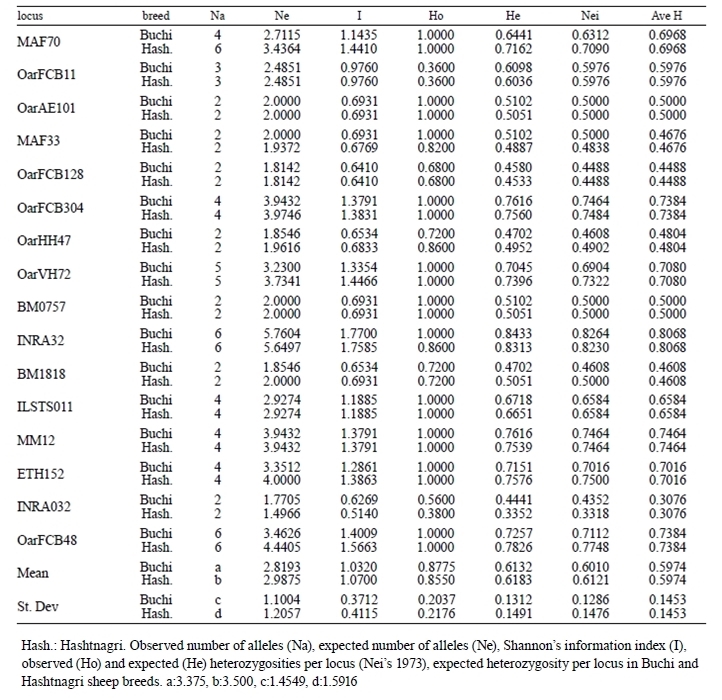
Number of alleles per locus ranged from 2 (Oar- AE101, MAF33, OarFCB128, OarHH47, BM1818 and INRA032) to 6 (MAF70, INRA32, ETH152 and OarFCB48) mean number of alleles (MNA) for Buchi and Hashtnagi populations were 3.375±1.455 and 3.500±1.592 respectively.
The observed heterozygosities in studied breeds ranged from 0.360 (OarFCB11) to 1.000 (MAF70, OarFCB304, OarVH72, ILSTS011, MM12, ETH152, OarFCB48) with an average of 0.878±0.204; while expected heterozygosities throughout all the breeds ranked between 0.458 (OarFCB128) and 0.843 (INRA32), and the mean was 0.613±0.131 for Buchi breed.
All the loci displayed higher observed heterozygosity for both breeds except OarFCB11. Mean Shannon information index (I) varied between 1.032±0.371 and 1.070±0.4112 for Buchi and Hashtnagi breeds respectively. The average PIC value for both of sheep populations were 0.53 (Buchi) and 0.59 (Hashtnagri). The FIS (inter-individual), FIT (total population) and FST (subpopulations) values tabulated on Table 3 where, only FST displayed positive values in terms of means. INRA32 and OarFCB11 markers were showed positive values on FIS for Buchi breed while only OarFCB11 showed positive values in Hashtnagi.
Table 3. Summary of F-statistics and gene flow for all loci in Buchi and Hashtnagri sheep breeds.
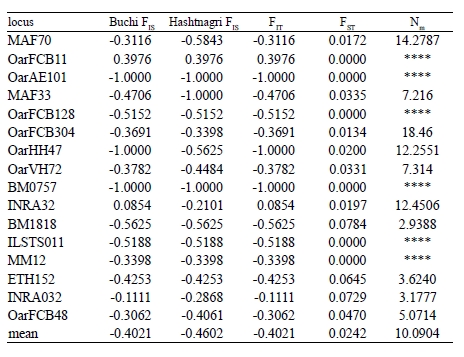
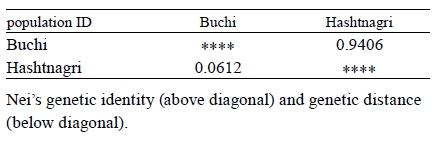
The mean FST value was 0.0242 reflecting the genetic differentiation estimates. Based on the FST values mean gene migration (or flow) (Nm) showed 10.09 between these breeds. The dendrogram based on Nei’s genetic distance using UPGMA method indicated the close genetic relationship between Buchi and Hashtnagri sheep breeds 43. Both sheep populations showed low genetic distance from each other (Figure 1) it was calculated as 0.0612 while genetic identity of Buchi and Hashtnagri sheep breeds was 0.9406 (Table 4) 43.
Figure 1. Dendrogram based Nei’s (1978). Genetic distance: method = UPGMA modified from Neighbor procedure of Phylip version 3.5.
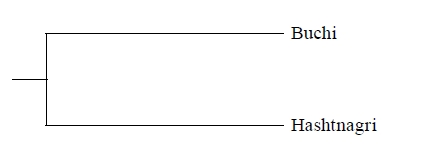
Table 4. Nei’s unbiased measures of genetic identity and genetic distance (Nei 1978).
DISCUSSION
A very limited study on genetic characterization of goat and sheep has been done in Pakistan, even though presence of more than 34 breeds of sheep and 30 breeds of goats has been reported 5, 37, 44. Most of these breed documentation has been carried out mainly based on morphological characters rather than the molecular basis. Genetic characterization is the foremost step towards utilizing the valuable animal resources effectively and efficiently. Moreover, genetic diversity studies have significant role in constructing the breeding strategies on economically important animal species 45.
Therefore, this effort is one of the important steps towards characterizing Pakistani sheep breeds. Most of the selected microsatellite markers showed polymorphism indicating their usefulness. All loci displayed at least two alleles which is a good indicator for the genetic variability among these breeds. Based on the number of alleles observed in all analyzed loci, considerable amount of genetic variability reported on these two breeds (>2) 46. Allelic richness is considered to play an effective role in conservation genetics due to the marker assisted selection methods for optimizing the number of alleles conserved in populations 47, 48.
The average heterozygosity of both Pakistani sheep breeds is lower than Chinese goat populations 0.677 to 0.737 49 and 0.77 to 0.821 50 and higher then some of the Indian goat and sheep breed values 0.426 and 0.501 9, 51. The observed heterozygosity in Buchi and Hashtnagri sheep (0.832, 0.877) breeds was higher than expected heterozygosity values of some other sheep breeds studied indicating no overall loss in heterozygosity (allele fixation) in the studied breeds 52 reports on gene diversities of Balkhi (0.547), Hashtnagri (0.533) and Michni (0.561) sheep populations of Pakistan, where this study reports slightly higher genetic diversity when compared to the Hashtnagri population.
Kail sheep population of Pakistan showed slightly lower numbers for heterozygosity measurements in comparison to the Buchi and Hashtnagri sheep populations reported here 53. Inbreeding measurements also linked to selection within and across breeds where, FST used to indicate subdivisions in population. A value of FST around 0.05 depicts moderate differentiation and <0.05 indicate lower differentiation between populations/ breeds 54. Mean values of FIS (heterozygote deficit) were -0.515 for goat breeds and -0.431 for sheep breeds are low representing low inbreeding coefficient between breeds which is obvious from the home track distance of the studied breeds in contrast to the positive FIS values (high level of inbreeding) in Italian sheep breeds 55.
Significant amount of inbreeding has been observed on sheep and goat diversity project in marginal areas carried out by Eu-Econogene project 56, 57. Generally marginal areas are characterized by small herds and improper breeding strategies lead to the high inbreeding, ultimately pave way to the fragmentation and isolation at the different breed levels 58. In general, low level of inbreeding (<0.10) can be a result of lower levels of mating between closely related individuals in field conditions 59.
Polymorphism information content (PIC) is another good indicator of the efficiency of marker for genetic studies. In this study the average PIC value of 16 loci was 0.53 in Buchi and 0.59 in Hashtnagri sheep which was lower than Indian domestic goat (0.60) 60, Chinese (0.62) and Saanen (0.57) goat breeds but greater than Korean goat (0.35) 61. The high average PIC values of panel of microsatellites used for all the breeds supported the appropriateness of the markers for genetic diversity analysis in the Pakistan sheep breeds, since the mean value is >0.5 62. However, allelic richness also depends on sample size and number of observed alleles. When the sample size is increased the presence of unique alleles within populations, which occurs in low frequencies, tends to increase.
In population diversity studies Shannon index is the most widely used which considers the abundance and evenness of the species studied 63. The Shannon index information also showed the mean value around one for both breeds, depicting the richness and also relatively low species richness and evenness of these breeds since the index showed less than 3.5 which is the mark set for high species evenness and richness 64. This might be due to the management system rather than the un-connected with the level of heterozygosity deficiency observed among the population.
From this study it can be concluded that Pakistani sheep breeds showed a high genetic diversity and the genotyping through microsatellites is an effective tool for the genetic evaluation of different breeds. This microsatellite panel can be used on other domestic and wild sheep breeds of Pakistan. The difference in the values of this study with other reports may be due to the difference of markers, breeding plans, laboratory techniques and sample size.
The results will help the future researchers as basic guide for better understanding of genetic relationships and breed differences in sheep and goat breeds for making future breeding policies and conservation plan to protect any loss in allelic variation in goat and sheep breeds in the country.
Acknowledgements. We would like to thank Higher Education Commission of Pakistan for funding the sheep and goat genetic diversity project (Nº 20-872) and Livestock and Dairy Development Departments of the Punjab and Khyber Pakhtunkhwa for helping in sample collection.
REFERENCIAS
References. El artículo incluye 64 citas bibliográficas que no se insertaron por su larga extensión. Los interesados pueden solicitarlas a faiz.marikar@fulbrightmail.org














