Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO  uBio
uBio
Compartilhar
Boletín de la Sociedad Argentina de Botánica
versão On-line ISSN 1851-2372
Bol. Soc. Argent. Bot. v.41 n.3-4 Córdoba ago./dez. 2006
Genotypic Diversity of the Wheat leaf blotch pathogen (Septoria tritici) in Buenos Aires Province
Cristina Alicia Cordo1, Celeste C. Linde2, Jashiu Zhan3 and Bruce Mcdonald3
1Research Scientist (CIC, Buenos Aires Province). CIDEFI-Facultad de Ciencias Agrarias y Forestales de La Plata. UNLP, calle 60 y 119. (1900) La Plata, Buenos Aires.
2School of Botany and Zoology, Bldg 116, Dalcy Rd, Australian National University, Canberra 0200, ACT Australia.
3Institute of Plant Sciences, Phytopathology Group, ETHZentrum/LFW, Universitatstrazsse 2, 8092 Zurich, Switzerland.
Summary: The population structure and genotypic diversity of Septoria tritici Rob ex Desm. from two crop field populations in Buenos Aires Province were studied with DNA restriction fragment length polymorphism. Hierarchical samples were taken from different cultivars. A total of 120 single spore isolates was subject to molecular analysis to compare the genetic structure of Los Hornos and Balcarce populations separated by 500km. Eight RFLP loci and 1DNA fingerprinting were used. Among the total of the isolates, 24 RFLP alleles were detected from the first population and 22 from the second. Gene and genotype diversity were high in both populations. Different haplotypes for each region were identified. Identical haplotypes were clustered in the same location in the field. The mean gene diversity and the mean genotypic diversity were high for the 8 loci of RFLP. This means that Balcarce and Los Hornos were significantly different. The X 2 was significant (P>0.005) only for two RFLP locus, then both populations were not independant. They had to be compared with an European and an American populations. Balcarce population was very different to all of them. Both Argentine populations were very similar at regional level but strongly different at a continental level. The greatest genetic variability between continents belonged to Argentine population. The amount of gene flow was high when all the populations were compared.
Key words: DNA hybridization; Septoria tritici; Mycosphaerella graminicola; RFLP; Epidemiology; Genetic diversity; Genotypic diversity.
Resumen: Diversidad genotípica del patógeno de la mancha de la hoja del trigo ( Septoria tritici ) en la provincia de Buenos Aires. Se estudió la estructura poblacional y la diversidad genética y genotípica de dos poblaciones de Septoria tritici Rob ex Desm. con la técnica de RFLP Un total de 120 aislamientos monospóricos colectada en campos experimentales de la Provincia de Buenos Aires se analizaron molecularmente, comparando la estructura genética de dos poblaciones separadas por 500 km . Se experimentó con 8 loci de RFLP y una sonda de ADN fingerprinting. Sobre la totalidad de los aislamientos se detectaron 24 alelos de RFLP para la localidad de Los Hornos y 22 para Balcarce. En la población de Los Hornos entre 58 aislamientos se reconocieron 35 haplotipos multilocus y en Balcarce, entre 62 aislamientos, 39 haplotipos, significando que ambas poblaciones son diferentes. Las diversidades genética y genotípica fueron altas en ambas poblaciones. La diferencia entre poblaciones, para las frecuencias de alelos, se calculó con el test de contingencia X 2 ; como estas fueron solo significativas para dos locus de RFLP (P>0.005) se procedió a comparar las poblaciones locales con una de europea y otra americana. Las poblaciones en Argentina fueron muy distintas a las otras poblaciones analizadas. Se interpretó que las poblaciones argentinas fueron similares a nivel regional pero difirieron significativamente de las de Suiza y Oregón, para los loci analizados.
Palabras clave: Hibridización de ADN; Septoria tritici; Mycosphaerella graminicola; RFLP; Epidemiología; Diversidad genética; Diversidad genotípica.
INTRODUCTION
Foliar fungal pathogens possess significant threats to grain crop production. Septoria diseases of wheat ( Mycosphaerella graminicola , anamorph Septoria tritici) cause economically significant yield losses in most of the wheat growing areas of the world. Losses can range from 31 to 54% in climates conductive to diseases development (Eyal et al., 1985). In Argentina, Annone et al. (1991, 1993) reported yield losses from 20 to 50% and Simón et al. (1996) found reductions in thousand kernel weights of 3 to 13%.
The primary genetic marker used for S. tritici in Argentina, was virulence (Cordo & Arriaga, 1990; Cordo et al., 1990/91; Giecco et al., 2004; Perelló et al., 1990). It was studied on a set of wheat differentials that varied in their level of resistance. However, virulence in this fungus is a character that is very sensitive to environmental conditions, originating difficulties to evaluate it in a reproducible manner
The population genetic structure of S. tritici has been characterized extensively by combining computer modeling (Zhan et al., 2004) experimental evolution approaches (Zhan et al., 1998, 2002) and population surveys based on restriction fragment length polymorphism (RFLP) (Zhan et al., 2001, 2003, 2004). Populations in this fungus are in genetic equilibrium as well as in drift migration equilibrium (Chen et al., 1996) attributed to a high rate of sexual recombination. Field populations sampled from different regions and continents shared similar RFLP frequencies suggesting that substantial gene flow has occurred across long distances (Zhan et al., 2003).
Genetic variations using restriction fragment length polymorphism (RFLP) began to be studied recently (Cordo et al. 1998, Cordo et al . 2006) in Argentina. Moreover the major wheat-growing regions of Buenos Aires Province have not been sampled extensively and it is not known whether results from other parts of the world are representative of these wheat growing regions. A set of genetic markers based on RFLP that could be used to estimate the amount and distribution of genetic variability, gene and genotype diversity, gene flow and the DNA fingerprints to identify clones. (Boerger et al., 1993; McDonald & Martínez, 1990a, 1990b, 1991; McDonald et al., 1995; Shaw & Royle, 1989) were developed in USA and Europe based on DNA radioactive labeling of the probe (McDonald & Martínez, 1990a).
The objectives of this work was to apply RFLP markers to compare the genetic structure of the Los Hornos and Balcarce populations that are 500 km distant from each other and to assess the potential for gene flow between both populations.
MATERIALS Y METHODS
The leaf infected tissues were collected from plants on GS10.1 stage (Zadocks et al. , 1974) in Los Hornos and Balcarce (Buenos Aires Province, Argentina) (Fig. 1). The hierarchical sampling method was used with each plant to collect S. tritici isolates from a naturally infected wheat field of Los Hornos. Fifty eight isolates originated from 39 lesions on 13 leaves were sampled at three different times in a single field. Each of them was approximately 10 m 2 in area (Table 1).

Fig. 1. Wheat productive areas in Buenos Aires province. X is indicating the place of sampling.
Table 1. Reference isolates of Septoria tritici from Los Hornos locality, Buenos Aires Province.

Table 1. Cont. aLos Hornos locality; bSTAR98 Septoria tritici Argentina year of collection 1998. c h Each digit in the haplotype corresponds to the allele present at each of the eight RFLP loci identified by a specific probe-enzyme combination.

A different strategy was used to sample a population of the Balcarce location. Collection was made from some experimental cultivars in the Agricultural Experimental Station, INTA. Cultivars that differed in resistance to S.tritici were planted in repetitions of 6 rows per cultivar. Thirty seven infected leaves in total, were randomly chosen from different cultivars. Sixty two isolates originated from 57 lesions were sampled. The total sampled area was 80 m 2 (Table 2). The infected wheat leaves were air-dried at room temperature for 2 wk before the fungi isolations were done. Only one single-spore isolate was obtained with the disinfections technique (alcohol 70% and Cl 2 Hg 1/1000 g/ml) and cultivated on PDA (2%) (Plant Pathologist's Pocketbook, 1968). The isolates were grown in yeast sucrose broth at room temperature (18-22 Cº) during 10 days with shaking (150 rpm) to obtain the spore concentration for further DNA extraction (McDonald & Martínez, 1990).
DNA was extracted from each isolate by a CTAB extraction protocol described previously (McDonald & Martínez, 1990; Cordo et al., 2006).
Anonymous probes for this study were selected using three criteria: 1) one repetitive probe to detect fingerprinting (pSTL70), 2) probes that produced a strong hybridization signal and hybridized to only one or two fragments (pSTL10, pSTL31, pSTS43, pSTS14) and 3) probes that hybridized to fragments between 0.5-6.0kb to insure adequate resolution of all restriction fragments present in our sample (pSTL53, pSTS192A, pSTS192B, pSTS2). All probes were handed over by Dr. McDonald. Purified DNA (5ug.) was digested individually with the PstI restriction enzyme. DNA fragments were separated on 0.75% agarose gels and then transferred to a nylon membrane by the alkaline transfer method (Reed & Man, 1985) as recommended by the manufacturer (BioRad, Hercules C.A.).
The plasmid was recovered from the Escherichia coli (HB101 strain) culture with the wizard Maxiprep DNA Purification System (Promega). Probes were labeled by radioactivity with P32, by nick translation, following the manufacturer's recommendations (BRL, Gaithersburg, MD). The single locus probes used for this experiment are shown in Table 4.
Each probe-enzyme combination defined an RFLP locus. DNA fragments or combinations of fragments of different sizes were treated as alleles at each RFLP locus. The number of isolates used in each analysis varied because of differences in the sampling methods for the two populations. Sample sizes of each locus also varied because data from some isolates were incomplete as a result of partial digestion, differences in the amount of DNA loaded in each line, and occasional nonspecific background hybridization. Only alleles that could be scored unambiguously were included in each analysis.
Genetic variation in each field population was quantified using measures of gene diversity (Nei, 1972). This measure of population differentiation was calculated to examine inter-population diversity among field populations. In addition a measure of genotypic diversity based on the number of multilocus haplotypes was calculated in each population. The total gene diversity was partitioned into several spatial components using hierarchical gene diversity analysis (Nei, 1972). Only the eight RFLP loci that were shared across all populations were included in the hierarchical analysis. Genotype diversity (G) in a population, based on the comparison of either multilocus haplotypes or DNA fingerprinting patterns combined with multilocus haplotypes, was calculated using the measurement proposed by Stoddart & Taylor (1988).The genetic similarity was assayed in several ways. Individual RFLP loci were compared directly in terms of allele frequencies, number of alleles per locus, Nei´s diversity, Neis´s genetic distance and identity. If two populations were not similar in size, then the gene flow (Nm) within each geographic region and among the regions was estimated with the method described by Nei (1972) who estimated the average number of individuals that migrate between the populations per generation. If Nm < 1 local populations will differentiate; if Nm> 1 there will be little differentiation among populations. Isolates with the same DNA fingerprints and multilocus haplotype were assumed to be individual members of the same clone and were counted only once in the analysis, as was demonstrated in previous experiments (McDonald & Martínez, 1991).To compare allele frequencies in the two populations, was used only one representative of each clone to calculate the clone corrected allele frequencies.
RESULTS
From of the 137 isolates coming from different areas of the Argentine wheat region, only 120 were characterized using the RFLP technique with P32 labeled probes. The pSTL70 fingerprinting probe hybridized many DNA fragments of different sizes in isolates from field populations of both places. All leaf samples were processed for isolation of the fungus, followed by fungus culture, DNA extraction, Pst1 enzyme digestion, radioactive hybridization and X ray film detection. Some of the isolates did not yield good quality DNA for the restriction enzyme digestion process. This explains the loss of 17 isolates in the samples of the populations.
In total, 24 alleles were found for Los Hornos population and 22 alleles for Balcarce population at the eight RFPL loci (Table 3). Despite the difference in the number of alleles, Nei´s measure of genetic diversity across all loci was different for both populations (0.2619 for Los Hornos and 0.3161 for Balcarce (Table 4).
Among the 58 isolates of Los Hornos and 62 of Balcarce (Table 2) with complete data from individual RFLP loci, 35 multilocus haplotypes for the first locality and 39 for the second locality, were registered. Seven new haplotypes (3a, 20a, 71a, 37a, 47a, 52a, 58a) were added to the list published on Internet ( Septoria tritici RFLP alleles, Official allele numbers and approximate fragment sizes as July 2000).The haplotype frequency in % (times that each haplotype is repeated over the total) varied from 1.72 (1 time) to 36.20 (21 times) for Los Hornos population and 1.61(1 time) to 14.50 (9 times) for Balcarce population (Table 1 and 2). Genotype diversity was greater in the Balcarce population (G=31.61 or 26.34% of the theoretical maximum of 120) than in the Los Hornos population (G=26.19 or 21.82% of the theoretical maximum of 120). On the other hand the mean genetic diversity between populations-Nei´s formula ( Ht = 0.405) was high for the 8 loci of RFLP (Table 5). This result implies that a significant difference exists between the populations of the two localities.
The result of the observation at a micro geographical level (ex. fungus isolated from 3 leaves of the same plant on the same place of collection) for Los Hornos (Table 1.-LHA30, LHA31, LHA33 or LH621, LH622, LH623·) and Balcarce (Table 2.-A31, A32,A33 or C41, C42) populations , demonstrated they had different genotypes of the pathogen when they were isolated from each of the three leaves; the same result was observed comparing leaves from different plants on the same place of collection (LH633, LH661 for Los Hornos or G23, G33 for Balcarce).
The isolates of Los Hornos and Balcarce populations showed a high number of fragments in the hybridization patterns with the pSTL70 probe. Fifty eight multilocus haplotypes and thirteen fingerprint patterns were registered for Los Hornos population and 55 multilocus haplotypes and fourteen fingerprint patterns for Balcarce population (Table 3). Many isolates of both populations had from one to several haplotypes for each fingerprint pattern. In Los Hornos population the E fingerprint pattern was present on 14 different haplotypes but it corresponded 3 times with the same 11211611 haplotype. In Balcarce the same fingerprint was present on 11different haplotypes, but it corresponded 8 times with the 11111211 haplotype. This last result is showing that there are clones in both populations. Some genotypes were detected as shared across the populations. In other cases several individuals in the two populations had the same multilocus haplotypes but different DNA fingerprints indicating that they were not the same clone.
The alleles frequencies (Table 4) were significantly different from the 8 loci of RFLP. Only for two RFLP locus the chi-square test was significant (P>0.005). Then the Argentine population must be compared with other continental populations -the Swiss and the United State (Oregon)- as independent populations. Estimates of Nei´s measures of gene diversity (Ht), total gene diversity (Hs) population differentiation (Gst), and gene flow (Nm) for all loci were summarized in Table 5. Over a total of 834 individuals, there was a 40% of gene diversity between native populations, and the total population differentiation was 11% showing that the differentiation between native and foreign populations exists. The low values of Gst indicate that these populations were virtually indistinguishable for these eight RFLP loci.
The average number of migrants (Nm) is 3.68. This number meant that 3 to 4 individuals would need to be exchanged across populations each generation to maintain the observed level of genetic similarity. Moreover the amount of gene flow between populations was high when all the populations were compared.
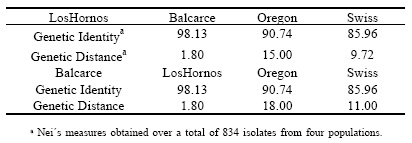
The analysis of the Genetic Identity and Genetic Distance between populations (Table 6) showed that the genetic distance was small when comparing the population of Los Hornos with the other populations, showing a high level of similarity, but the genetic distance of Los Hornos and Balcarce populations was mayor comparing with Oregon and Swiss populations. Salamati et al . (2000) suggested that the similarity among populations on a regional basis was explained because the gene flow was significant over spatial scales of at least several hundred kilometers. It found that genetic distances among fields within a region were small, while genetic distances among different continents were larger for the Rhynchosporium secale populations.
Table 2. Reference isolates of Septoria tritici from Balcarce, Buenos Aires Province.


Table 3. Information summary for two populations of Septoria tritici from Argentina.

Table 4. Clone- corrected allele frequencies, Nei´s measures of gene diversity for eight restriction fragment length polymorphism loci in two field populations of Septoria tritici from Buenos Aires Province
Table 5. Population structure and gene flow among Los Hornos, Balcarce, Oregon and Swiss populations.

aAll alleles are based on the digestion of DNA with PstI, b Sample size, c gene diversity among populations, d total gene diversity, e population differentiation, f gene flow.
Table 6. Table 6. Nei´s measures (in %) of Genetic Identity and Genetic Distance among Septoria tritici populations from Los Hornos and Balcarce (Argentina), Oregon (USA) and Switzerland.
DISCUSSION
RFLP markers used in this study revealed a high degree of genetic variability in the two Argentine populations. Higher genotype diversity was observed in the population from Balcarce than in Los Hornos. This would be likely do to the different sampling strategy used to collect the isolates from Los Hornos. The shorter distance between sampling points, choose on a hierarchical sampling, increased the likelihood of finding the same clone resulting from splash-dispersal of conidia as was demonstrated by Salamati et al. (2000). In coincidence with the same author the analysis of the local populations shown that the genetic distance among field populations were relatively small within continents and larger between continents. The complete Balcarce sample included 62 isolates from 57 leaves practically, each isolate coming from a different leaf; on the other hand, Los Hornos sample had 58 isolates coming from 40 leaves of different wheat varieties and different times of collection. Both populations shared the same alleles; different sites from the same population also repeated the same alleles. Most of the additional alleles present in the Balcarce population occurred at a low frequency, often in only one individual.
The genotypic diversity of the Balcarce population, which was 31.61% , was higher than that of the Los Hornos population, which was only 26.19% . Most of the isolates in the Los Hornos sample with the same multilocus haplotype were isolated from the same leaf. Genotypic diversity within populations and similarity over regional spatial scale was explained because regular sexual recombination was occurring rather in Septoria tritici than in Rynchosporium secalis (Salamati et al., 2000), Stagonospora nodorum (Mc Donald et al., 1994) and Phaeosphaeria nodorum (Keller et al., 1997) populations. This was explained because the ascospores from the teleomorph were dispersed over distances of up to hundred of kilometers (Shaw & Royle, 1989; Cordo et al ., 1990/1991; Cordo et al. , 2005).
In the Balcarce field, each multilocus haplotype was repeated from one to twenty one times maximum; but in Los Hornos more than one multilocus haplotype was repeated several times in coincidence with the same DNA fingerprint. These haplotypes were considered clones and it existence was confirmed by the low frequency of the mating type MAT1.2 associated to that locality (Albani et al., 2005). They appeared because Los Hornos are not an endemic area for the leaf blotch of wheat and the varieties that are checked for resistance are artificially inoculated with the pathogen, then the gene diversity is low in this field. In coincidence with McDonald et al. (1998) and Jurgens et al. (2006), these isolates could be individuals that were not sexually compatible to create major diversity, for climatic or other adversities, leading to genetic disequilibrium.
The field populations of the fungus exhibited high degrees of gene and genotype diversity distributed on very small spatial scales. The micro geographical level observations showed a higher variation on type and number of genotypes for the Balcarce than for the Los Hornos population. In general, different genotypes were often found within a single lesion, and most lesions on the same leaf had also different genotypes. This result demonstrated, in coincidence with Boerger et al. (1993), that a lesion is often the result of a co infection by two or more genotypes.
The genetic distance, for native populations, were very small, considering that the geographic distances between them was 500km; the North American and European populations, separated by to 7000km, had a low increase of this genetic distance. Then the high degree of similarity could be caused by the gene flow on a regional scale and between continents (Banke et al., 2004; Banke & McDonald, 2005; Boerger et al., 1993; Zhan et al ., 2003).
The results of this contribution are in agree with Keller et al. (1997) who demonstrated that ascospores are the primary agent for unifying geographically separated populations on a regional scale. Added to this, Cordo et al. (2005) showed that ascospores were the most significant component of the Mycosphaerella graminicola life cycle in the wheat producing areas in Argentina. Their release was registered in the vegetative and debris wheat states for the analyzed periods. According to these experience the high degree of gene flow among populations would be associated neither the pycnidiospores presence as dominant in the life cycle of the pathogen nor the infected seeds that could act as human dispersal mechanism (Keller et al., 1997). Los Hornos population resulted different because the lineages clonal of Septoria tritici were probably originated from the inoculations applied for the resistance tests.
If it is assumed that Septoria tritici did not recently colonize Argentine, the high degree of similarity could be explained from the most likely center of origin for this pathogen. Banke et al., (2004) demonstrated that the New World areas (where the South Cone is considered), appeared less likely to represent ancestral populations because they had lower diversity, whereas Israel and Europe appeared to be the ancestral populations because they shown the highest genetic diversity. This pattern was related with the fact that the wheat has been grown in the Old World for thousands of years, but in the New World for only hundreds years. The movement of the fungus from Israel and into Europe could have been from wind blown ascospores or via transport on infected seed or straw. Ascospores movement produced a natural gene flow out of the possible center of origin and into European populations that could explained the finding that more haplotypes were found in European than in New World populations.
At least, another way of dispersion could be an alternate host of S. tritici , producing pycnidia, that constitute a continuous host population where ascospores (Boerger et al. , 1993; Linde et al., 2002) would maintained a uniform source of inoculum that infects the wheat field each autumn This way of transmission was not demonstrated in Argentina.

Fig.2- Examples of DNA fingerprints and RFLPs in nuclear DNA of Septoria tritici isolates sampled from a Balcarce field. The same isolates are shown in each panel. All DNA was digested with PstI. (A) Probe pSTL70 hybridized to a dispersed repetitive DNA family. (B) Probe pSTL53 hybridized to two loci located on different chromosomes. The upper bands represent alleles at one locus and the lower bands represent alleles at a second locus. (C ) Probe pSTL10 hybridized to one locus. ( D) Probe pSTS 2 hybridized to one locus.
CONCLUSIONS
We used DNA restriction fragment polymorphism (RFLP) markers labeled with radioactive compounds to assess the potential for gene and genetic diversity and for gene flow between geographically separated populations.
The results about the genetic compositions of two populations separated by 500 km are showing shared haplotypes. This has significant implications for wheat breeding programs that seek to incorporate resistance to S. tritici . In coincidence with Boerger et al . (1993), our evidences for gene flow suggest that plant breeders in Argentina are driving the breeding process well. They are testing the resistance of their cultivars at many locations away from the area of local adaptation. The fine scale of patterns with genetic variability suggests that plant breeders should use a wide spectrum of pathogen genotypes when testing wheat resistant cultivars to this pathogen in any location.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the assistance of Ings. Agrs. J. Bariffi, Juan Annone, Gilberto Kraam, and Lic. Natalia Kripelz, who collected much of the infected leaf tissue from which the isolates used in this study originated. There is a special acknowledgment to Dr. B. McDonald and his group for guide me on this subject. This project was supported by CIC Exps. 2157- 719/03, 2157-125/04 and CONICET PICT 99 (08-06356ªA).
BIBLIOGRAPHY
ALBANI C. M., F. CONSOLO, G. SALERNO & C. CORDO. 2005. Determinación de alelos de compatibilidad sexual en poblaciones de Mycosphaerella graminicola de Argentina. En Libro de Resúmenes XIII Congreso Latinoamericano de Fitopatología, III Taller de la Asociación Argentina de Fitopatología: 339. 19-22 de abril de 2005. Villa Carlos Paz, Córdoba. [ Links ]
ANNONE, J., A. CALZOLARI, O. POLIDORO & H. CONTA. 1991 Efecto de la mancha de la hoja causada por Septoria tritici sobre el rendimiento. INTA EEA Pergamino. Informe Nº122, 4. [ Links ]
ANNONE, J., H. CONTA, O. POLIDORO & A. CALZOLARI. 1993. información adicional sobre el efecto de 'mancha de la hoja´ causada por Septoria tritici sobre los rendimientos. INTA EEA Pergamino. Informe N 146, 5. [ Links ]
BANKE, S., A. PESCHON & B. A. MCDONALD. 2004. Phylogenetic analysis of globally distributed Mycosphaerella graminicola populations based on three DNA sequence loci. Fungal Genet. Biol. 41: 226-238. [ Links ]
BANKE, S. & B. A. MCDONALD. 2005. Migration patterns among populations of the pathogenic fungus Mycosphaerella graminicola . Molec. Ecol. 14:1881-1896 [ Links ]
BOERGER, J. M, R. S. CHEN & B. A. MCDONALD. 1993. Gene flow between geographic populations of Mycosphaerella graminicola (Anamorph Septoria tritici ) detected with restriction fragment length polymorphism markers. Phytopatology 83: 1147 - 1154. [ Links ]
CORDO, C. A. & H. O. ARRIAGA. 1990. Variación en patogenicidad entre cepas argentinas de Mycosphaerella graminicola (anamorfo Septoria tritici ). In: Kohli M. M. & van Beuningen L.T. (eds.), Conferencia Regional sobre septoriosis del trigo , pp. 88 -100. CIMMYT, México, D. F. [ Links ]
CORDO, C. A., A. E. PERELLÓ, H. E. ALIPPI & H. O. ARRIAGA. 1990/91 Presencia de Mycosphaerella graminicola (Fuckel) Schroeter telomorfo de Septoria tritici Rob. apud Desm. en trigos maduros de la Argentina. Rev. Fac. Agron . 66/67: 49-55. [ Links ]
CORDO, C. A. & M. M., LOJO. 1998. Estudio del polimorfismo en la longitud de fragmentos de restricción (RFLP) de aislamientos de Mycosphaerella graminicola mediante el uso de sondas no radioactivas. Fitopatologia 31: 22. [ Links ]
CORDO, C. A., M. R. SIMÓN, A. E. PERELLÓ, D. BAYO, N. KRIPELZ & S. LARRÁN . 2005. Environmental factors affecting the release and dispersal of pycnidiospores and ascospores of Mycosphaerella graminicola. . 7 th International Wheat Conference: 131. Mar del Plata. [ Links ]
CORDO C.A., M.M. LOJO, P. REMORINI. 2006. Two Adapted Techniques in Studies of DNA Fingerprinting of Septoria tritici Populations. Plant Pathol. J., 5: 41-46. [ Links ]
CHEN, R.S., M. BOERGER & B.A. MCDONALD. 1994. Genetic stability in a population of a plant pathogen in fungus over time. Molec. Ecol . 3: 209-218. [ Links ]
CHEN, R. S. & MC DONALD, B.A. 1996. Reproduction Plays a Major Role in the Genetic Structure of Populations of the Fungus Mycosphaerella graminicola. Genetics. 142: 1119-1127. [ Links ]
EYAL, Z., SCHAREN, M.D., HUFFMAN, M.D., AND PRESCOTT, J.M. 1985. Global insights into virulence frequencies of Mycosphaerella graminicola. Phytopathology 75: 1456-1462. [ Links ]
GIECO, J. O., J. DUBCOVSKY & L. E. ARANHA CAMARGO. 2004. Aggressiveness and physiological specialization of Septoria tritic i isolates. Sci. Agriac. (Piracicaba, Braz .) 61: 414-421. [ Links ]
JÜRGENS, T., C. LINDE, B. MCDONALD. 2006. Genetic structure of Mycosphaerella graminicola populations from Iran, Argentina, and Australia. Europ. J. Plant Pathol . On line version. Springer, Berlin. [ Links ]
KELLER, S. M., M. S. WOLFE, J. M. MCDEMOTT & B. A. MCDONALD. 1997. High genetic similarity among populations of Phaeosphaeria nodorum across wheat cultivars and regions in Switzerland. Phytopathology 87: 1134-1139 [ Links ]
LINDE, C., J. ZHAN & B. A. MCDONALD. 2002. Population structure of Mycosphaerella graminicola : from lesions to continents. Phytopathology 92: 946-955. [ Links ]
MCDONALD, B. A. & J. P. MARTINEZ. 1990a. DNA restriction fragment length polymorphism among Mycosphaerella graminicola (anamorph Septoria tritici ) isolates collected from a single wheat field. Phytopathology 80: 1368-1373. [ Links ]
MCDONALD, B. A. & J.P. MARTINEZ. 1990b. Restriction fragment length polymorphisms in Septoria tritici occur at a high frequency. Curr. Gen. : 17: 133-138. [ Links ]
MCDONALD, B.A. & J.P. MARTINEZ. 1991. Chromosome length polymorphisms in Septoria tritici population. Curr. Gen . 19: 265-271. [ Links ]
MCDONALD, B. A., J. MILES, L. R NELSON, R. E. PETTWAY. 1994. Genetic variability in nuclear DNA in field populations of Stagonospora nodorum. Phytopathology 84: 250-255. [ Links ]
MCDONALD, B. A., R. E. PETTWAY, R. S. CHEN, J. M BOERGER & J. P. MARTINEZ, 1995. The population genetics of Septoria tritici (telemorph Mycosphaerella graminicola ). Canad. J. Bot. 73 (Suppl.): S292-S301. [ Links ]
MCDONALD, B. A., J. ZHAN, K. YARDEN, J. HOGAN, J. GARDON, & R. E. PETTWAY. 1998. The population genetics of Mycosphaerella graminicola and Stagonospora nodorum. In: Understanding pathosystem: a focus of Septoria. 15 th Long Ashton International Symposium IACR-Long Ashton research Station, Bristol, England Chapter 3: 44-69. [ Links ]
NEI, M. 1972. Genetic distance between populations. Amer. Nat . 106: 283-292. [ Links ]
PERELLÓ, A., F. J. BABINEC & CORDO, C. A. 1990. Especialización fisiológica en cepas argentinas de Mycosphaerella graminicola (Fuckel) Schroeter (Anamorfo). Septoria tritici Rob. ex Desm. In: Kohli, M. M. & van Beuningen L. T. (eds), Conferencia Regional sobre Septoriosis del trigo , pp. 101-107. CIMMYT, México, D.F. [ Links ]
PERELLO, A., C. A. CORDO, H. O .ARRIAGA. & H. E. ALIPPI. 1991. Variation in virulence in isolates of Septoria tritici Rob. ex Desm. on wheat. Agronomie 11: 571-579. [ Links ]
PLANT PATHOLOGYST´S POCKETBOOK. 1980. The Commonwealth Mycological Institute. C. A. B. The Commonwealth Mycological Institute: 267. Kew, Surrey. [ Links ]
REED, C. K. & D. A. MANN. 1985. Rapid transfer of DNA from agarose gel to nylon membranes. Nuclei Acids Res . 13: 7207-7221. [ Links ]
SHAW, M. W. & D. J. ROYLE. 1989. Airborne inoculum as a major source of Septoria tritici ( Mycosphaerella graminicola) infections in winter wheat crop in the U.K. Plant Pathol. 38: 35-43 [ Links ]
SALAMATI, S., J. ZHAN, J. J. BURDON, B. MC DONALD. 2000. The genetic structure of field populations of Rynchosporium secalis from three continents suggests moderate gene flow and regular recombination. Phytopathology 90: 901-908. [ Links ]
SIMON, M. R., A. E. PERELLÓ & C. A. CORDO. 1996. Influencia de infección tardía de Septoria tritici Rob. ex Desm. sobre el peso de 1000 granos y algunos parámetros de calidad en Triticum aestivum . Investigación Agraria, Producción y Protección Vegetales 11: 162- 171, España. [ Links ]
STODDART, J. A. & J. F. TAYLOR. 1988. Genotypic diversity: estimation and prediction in samples . Genetics 118: 705-711. [ Links ]
ZADOCKS, J. C.,T. T. CHANG & C. F. KONZAK. 1974. A decimal code for the growth stages of cereals, Weed Res . 14: 415-421. [ Links ]
ZHAN, J.,C. C. MUNDT, M. E. HOFFER & B. A. MCDONALD. 1998. Measuring immigration and sexual reproduction in field populations of Mycosphaerella graminicola. Phytopathology 88: 1330-1337. [ Links ]
ZHAN, J., C. C. MUNDT & B. A. MCDONALD. 2001. Using restriction fragment length polymorphisms to assess temporal variation and estimate the number of ascospores that initiate epidemics in field populations of Mycosphaerella graminicola. Phytopathology 91: 1011-1017. [ Links ]
ZHAN, J., C. C. MUNDT, M. E. HOFFER & B. A. MCDONALD. 2002. Local adaptation and effect of host genotype on the evolution of pathogens: An experimental test in a plant pathosystem . J. Evol. Biol 15: 634-647. [ Links ]
ZHAN , J., R. E. PETTWAY & B. A. MC DONALD. 2003. The global genetic structure of the wheat pathogen Mycosphaerella graminicola is characterized by high nuclear diversity, low mitochondrial diversity, regular recombination, and gene flow. Fungal Genet. Biol. 38: 286-297. [ Links ]
ZHAN, J., G. H. J. KEMA & B. A. MCDONALD. 2004. Evidence for Natural Selection Mitochondrial Genome of Mycosphaerella graminicola. Phytopathology., 94: 261-267. [ Links ]
Recibido el 27 de Junio de 2006
Aceptado el 10 de Noviembre de 2006.














