Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
Boletín de la Sociedad Argentina de Botánica
versión On-line ISSN 1851-2372
Bol. Soc. Argent. Bot. vol.46 no.3-4 Córdoba jul./dic. 2011
GENÉTICA Y EVOLUCIÓN
Genetic diversity in a natural population of the halophytic legume Prosopis strombulifera revealed by AFLP fingerprinting
Analia Llanes1, Victoria Bonercarrere2, Fabian Capdevielle2, Sabina Vidal3 and Virginia Luna1,4
1Laboratorio de Fisiología Vegetal, Departamento de Ciencias Naturales, Fac. de Cs. Exactas, Físico-Químicas y Naturales, Universidad Nacional de Río Cuarto, Ruta 36 Km 601, 5800 Río Cuarto, Argentina.
2Instituto Nacional de Tecnología Agropecuaria. INIA-Las Brujas Ruta 48 Km 10, Rincón del Colorado, 90000 Canelones, Uruguay.
3Laboratorio de Biología Molecular Vegetal, Fac. de Ciencias, Universidad de la República, Iguá 4225, 11400 Montevideo, Uruguay.
4Corresponding author: vluna@exa.unrc.edu.ar, TE: +54-358-4676103. FAX: +54-358-4676230.
Summary: Prosopis strombulifera (Lam.) Benth. is a spiny shrub with the maximum tolerance limits reported for halophytic plants. This species is frequently found in the salinized areas in south-western of Córdoba and San Luis provinces, Argentina. Little is known about the genetic diversity within this species in a native population. Genetic diversity in 60 plants of P. strombulifera in south-western San Luis was investigated using AFLP analysis. Polymorphism was found among the samples tested. Four combinations of primers led to the identification of an average of 250 polymorphic bands and the data were used for cluster analysis. P. strombulifera genotypes are clearly separated in subclusters and reflect the diversity within the collection area. This study is a contribution to describe the intra-specific diversity in a natural population of P. strombulifera, and the polymorphism obtained is comparable with other populations of Prosopis species. Results demonstrate the importance of identifying different intra-population genotypes as components of a gene bank of P. strombulifera.
Key words: Prosopis strombulifera; AFLP; Genetic diversity; Polymorphism.
Resumen: Diversidad genética en una población natural de la leguminosa halófita Prosopis strombulifera revelado por el análisis de AFLP. Prosopis strombulifera (Lam.) Benth. es un subarbusto espinoso con limites máximos de tolerancia informado para especies halófitas. P. strombulifera se encuentra en los suelos salinizados del Sur-Oeste de las provincias de Córdoba y San Luis, Argentina. El conocimiento sobre la diversidad genética en poblaciones nativas de esta especie es escaso. En este trabajo se investigó la diversidad genética en 60 plantas de P. strombulifera mediante el análisis por AFLP. Se observó polimorfismo entre las muestras analizadas, cuatro combinaciones de cebadores identificaron un promedio de 250 bandas polimorficas, las que fueron utilizadas para análisis de agrupamientos. Los genotipos de P. strombulifera fueron separados en subgrupos reflejando la diversidad dentro del área de muestreo. Este estudio contribuye a describir la diversidad intra-específica en una población natural de P. strombulifera, y el polimorfismo obtenido es comparable al observado en otras poblaciones en especies de Prosopis. Estos resultados demuestran la importancia de identificar diferentes genotipos dentro de la población como componentes de un banco de genes de P. strombulifera.
Palabras clave: Prosopis strombulifera; AFLP; Diversidad genética; Polimorfismo.
Introduction
The genus Prosopis L. emend. Burkart belongs to Fabaceae (Leguminosae), sub-family Mimosoideae (Pasiecznik et al., 2001). The history of taxonomic confusion within the genus was largely settled with the authoritative monograph of Burkart (1976), who defined the generic limits and divided the genus in five sections with marked vegetative differences in armature (Pasiecznik et al., 2001).
While the generic limits and division into sections defined by Burkart (1976) are generally accepted, there is continuing debate as to the relative rank of species that he defined.
Prosopis includes about 45 species grouped in five sections, and has a wide distribution, occurring in South, North and Central America, Africa and Western Asia (Burkart, 1976; Schimi, 1981). Most of the species are concentrated in South America; Argentina is the center of greatest diversity with about 27 species, 11 of which are endemic (Burkart, 1976). This genus has been studied with growing interest during the last few years because Prosopis species have been widely introduced in several countries around the world due to their ability to grow in the poorest soils and survive in areas where no other species can survive (Golubov et al., 2001; Pasiecznik et al., 2001).
Prosopis strombulifera (Lam.) Benth. (Burkart, 1976) is a legume frequently found in the salinized areas of south-western San Luis in Argentina (Sosa, 2005). This halophytic shrub belongs to section Strombocarpa and the genetic divergence among its species has been addressed by Saidman et al. (1996). It ranges from the Arizona desert (USA) to Patagonia (Argentina), and is particularly abundant in the salinized areas of Central Argentina (Cantero et al., 1996). In these areas, proportions of NaCl and Na2SO4 are generally similar, although Na2SO4 is as much as three times more abundant in some samples (Sosa et al., 2005). It is important to compare effects of Na2SO4 and NaCl on plant growth, to better understand plant responses to the major salts found in salinized soils in various countries (Sosa et al., 2005; Manivannan et al., 2008). P. strombulifera can survive in hydroponic cultures containing as much as 1M NaCl with metabolic (enzymatic) changes in roots and leaves. Its seedlings showed a halophytic response to NaCl, i.e. growth of stems and roots increased as substrate-NaCl increased to 450-500 mM. Treated plants were more vigorous, had a higher leaf number with smaller leaflets, and harder and darker spines than the controls. Growth was slowly inhibited when NaCl concentrations exceeded 500 mM (Reinoso et al., 2004, 2005; Sosa, 2005). These results indicate that P. strombulifera is within the maximum tolerance limits reported for halophytic plants being much more tolerant than most Prosopis species and reaching the NaCl tolerance level of Chenopodium rubrum (Egan & Ungar, 1998). However, P. strombulifera was much less tolerant to Na2SO4 showing a strong general growth inhibition, when iso-osmotic solutions of this salt were compared to NaCl (Reginato, 2009). However, great physiological variability was observed in response to salinity under both, NaCl and Na2SO4 treatments. In previous studies from this group, determinations of growth parameters as well as ion concentration, hormone levels (Reginato, 2009; Llanes, 2010), total aminoacids, polyalcohols, and protein content (Llanes et al., 2011) showed high variability independently of the maintenance of the same experimental conditions. This could be reflecting the degree of intraspecific genetic variability in this species. Knowing the inherent genetic variability in a native population will facilitate the understanding of the observed variability in physiological responses and may help to identify tolerant or sensitive genotypes. In addition, knowledge of the genetic structure of native populations and the characterization of genetic variation are dominant factors in defining effective strategies and programs to improve understanding of the consequences of handling this variability (López et al., 2001).
Among the different marker systems available, the amplified fragment length polymorphism (AFLP) technique, first described by Vos et al. (1995), exploits the advantages of technical simplicity and generation of large numbers of markers spanning the whole genome without any prior knowledge about it. These markers are particularly suitable for genotypic evaluation of species like Prosopis since they are highly reproducible with overall error rates of less than 2%, amenable to automation for high throughput genotyping, and anonymous, so they do not require any sequence information. AFLPs have been successfully used to determine genetic diversity in many plant species, mostly crop science, and to construct genetic maps (barley: Becker et al., 1995; Waugh et al., 1997; melon: Wang et al., 1997, Arabidopsis; Alonso-Blanco et al., 1998).
The use of AFLPs is particularly well-suited when there is no a priori sequence information for intra-specific studies, when genomic heterogeneity is high (i.e. when it is necessary to amplify many loci to ascertain an accurate measure of genomic diversity, e.g. outcrossing species), and when genetic variability is low (i.e. when it is necessary to amplify many loci to locate the few that are polymorphic, e.g. crop species) (Bensch & Akesson, 2005; Meudt & Clarke, 2007).
In this study, genetic diversity within a natural population of P. strombulifera in the south-western of Córdoba and San Luis was investigated by using the AFLP technique, to allow a better understanding of the variability in the physiological responses to salt treatments observed in previous studies, in order to identify tolerant and sensitive genotypes.
Materials and Methods
Plant material. Seeds of P. strombulifera were collected in south-western San Luis, Argentina, at 33º43'S, 66º37'W and 400-500m of altitude, with a temperature thermal regime (i.e., an annual average temperature of 15-20ºC) (Fig. 1). This area belongs to the mesquite tree forest located in a saline depression between the annual 300 to 400 mm isohyets in the Monte phytogeographic region (Anderson et al., 1970; Peña Zubiate et al., 1988; Carosio et al., 2009). The soil has a franc sandy texture, with abundant calcareous material and moderate salinity (8000 mho/cm2 electrical conductivity at the surface). Pods were collected at random from 100 plants within the same population. About 100 legumes from several shrubs were collected along a transect of about 100 m. Approximately 70 seeds (individuals) were selected visually for uniform size and healthy aspect. They were scarified with sulphuric acid for 10 min and then washed overnight under flowing water. Before sowing they were rinsed in distilled water and placed in Petri dishes with two layers of water saturated filter papers at 37ºC for 24 h (Reinoso et al., 2004). Germinated seeds with 20 mm long radicles were cultured in hydroponic condition in two black trays per treatment per experiment (200 seedlings per tray) with 10% of full strength Hoagland's solution. The seedlings were grown in a growth chamber under a 16 h light (200 µmol.m-2.s-1) (28ºC) 8 h dark (20ºC) cycle and 70% relative humidity. After one week, seedlings were transferred to a 25% full strength Hoagland's solution (osmotic potential-0.11 MPa). The pH of the medium was 6 in all cases; aquarium tubing with a peristaltic pump system was used to provide aeration. DNA from 21 days old plants were used for AFLP analysis.

Fig. 1. Map of San Luis province indicating collection area of Prosopis strombulifera.
Genomic DNA extraction. Genomic DNA was extracted essentially as described by Dellaporta et al. (1983) with an additional RNase treatment and phenol extraction using plant material collected from 60 individuals. This material was homogenized adding 15 ml of extraction buffer (100 mM Tris-HCl, pH 8.0, 50 mM EDTA, pH 8.0, 500 mM NaCl , 10 mM B-mercaptoethanol), and 1 ml of 20% SDS; following shaking incubation at 65ºC for 15 minutes, samples were precipitated using 5 ml of 5 M KAc at 0°C for 30 minutes. The samples were centrifuged 15 minutes at 17000 rpm and 10 ml of isopropanol was added to supernatant, mixed and incubated at 20°C for 30 minutes. The mix was centrifuged 15 minutes at 17000 rpm and the pellet was washed with 70% ethanol and allowed to dry at room temperature. The pellet was resuspended in 450 µl TE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, pP 8.0) 50 µl of 3 M NaCl and incubated at 37°C for 30 minutes with 2 µl of RNase (10 mg/ml). The solution was transferred to an Eppendorf tube and the extract was washed with 1 vol of phenol and twice with 1 vol of chloroform. DNA was precipitated with 0.8 vol. isopropanol and was centrifuged for 1 min. The pellet was washed with 70% ethanol and was dried and suspended in 200 µl of TE buffer and was reserved at -20º C until further use.
AFLP Protocols. 1-Template Preparation and Adaptor Ligation. AFLP analysis was performed according to Vos et al. (1995) using AFLP Small Plant Genome protocol (PE Applied Biosystem, Foster City, CA). This protocol was adapted with the following modifications. Genomic DNA (500 ng) was digested with 10 U EcoRI (Fermentas Life Science) and 10 U MSeI (Fermentas Life Sciences) during 3 h at 37ºC. Double restriction reaction was carried out in 25 µl reaction volumes in Tango TM Buffer (Fermentas Life Science) containing 33 mM Tris-Acetate (pH 7.9), 10 mM MgAc, 66 mM KAc and 0.1 mg/ml BSA. EcoRI and MseI adapters were ligated to DNA with 5U of T4 Ligasa (Fermentas Life Science) in 20 µl reaction volume containing 40 mM Tris_HCL, 10 mM MgCL2, 10 mM DTT and 0.5 mM ATP (pH 7.8). The ligation was incubated overnight at 16ºC. After ligation, samples were diluted 10 fold.
2- Preselective amplification. This reaction was performed by adding 1 µl of Primer Mix and 15 µl of AFLP Core Mix to 4 µl of restricted-ligated-diluted DNA. The Primer Mix contains EcoRI and MSeI primers complementary to the adapters sequence with and additional 3´base following the parameters: 1 cycle of 2 min at 72ºC, 20 cycles of 30s at 94ºC, 1 min at 56ºC, 2 min at 72ºC and 1 cycle of 30 min at 60ºC. After PCR, samples were diluted 20 folds.
3- Selective amplification. Diluted-amplified DNA was used as template for selective amplification reactions. The reaction mixture contains 15 µl of AFLP Core Mix, 1 µl of each selective primer and 3.5 µl of diluted preselective amplification product. Only primers complementary to EcoRI adapters were labeled. The EcoRI and MSeI adapters, Preselective Amplification Primer Mix, AFLP Core Mix and primers for selective amplifications are supplied in AFLP Ligation and Preselective and Selective Amplification Module (PE Applied Biosystem, Foster City, CA). Four primer combinations were used in this study. Selective amplifications were done with different combinations of fluorescent labeled EcoRI primers with three selective bases and MseI primers with two or three selective bases. AFLP primer combinations used to characterize the genetic diversity were: EcoRI AAC-MSeI CAA, EcoRI AGG-MSeI- CTA, EcoRI ACC-MSeI CTC, EcoRI ACG-MSeI CTT.
4- Capillary electrophoresis. AFLP amplification products were separated by capillary electrophoresis using the FAM-JOE-TAMRA module. Selective amplification products (1-2 µl) were mixed with 11.5 µl Hi-DiTMformamide and 0.5 µl of DNA labeled with ROX dye (GeneScan-500 ROX PE Applied Biosystem, Foster City, CA). The samples were denatured at 95ºC for 5 min, kept on ice for 5 min and loaded onto the automated ABIPRISM 310 Genetic Analyzed. The run parameters were: injection time 5 to 10 sec, electrophoresis voltage 15 kv, collection time 25 min and temperature 60ºC.
5- Data analysis. Row data were analyzed using the Genotyper 3.7 Analysis Software. AFLP bands were scored as present (1) and absent (0) and only bands showing unambiguous polymorphism were entered into a data matrix that was used for the following analyses.
Genetic diversity among samples was calculated with the simple matching coefficient, followed by hierarchical clustering analysis using Ward method. Alternatively, a non hierarchical clustering approach (simple k-means, implemented within the Waikato Environment Knowledge Analysis -WEKA-) was used to estimate the likelihood of diverse cluster arrangements (Witten & Frank, 2005).
Results
Each of the four primer combinations was able to generate an informative AFLP fingerprint across all samples. Among the 60 genotypes studies, the four pairs of primers identified 251 polymorphic bands (Table 1).
Table 1. The primer combinations used for an amplified fragment length polymorphism (AFLP) analysis and the respective number of DNA fragments generated.

The genomic diversity explored using AFLP was evident both in the composition, and the number and frequency of the fragments analyzed. The number of markers scored per primer combination was 82. Each primer pair produced between 49 and 111 distinguishable AFLPs bands with an average of 63 per primer pair.
A hierarchical clustering dendrogram representation was constructed from similarity data (Fig. 2); the estimated co-phenetic coefficient (r) was 0.82, pointing towards a genetic diversification among diverse clusters. Under the non hierarchical clustering approach, within-cluster sum of squared errors was estimated (Fig. 3), pointing towards a heterogeneous structure, with some larger clusters (19, 16, and 14 accessions respectively) and some smaller clusters (3, 6 and 2 accessions respectively).
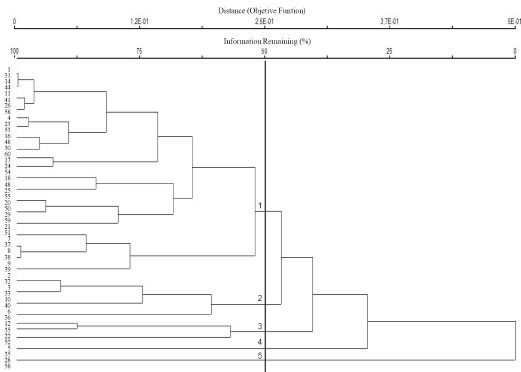
Fig. 2. Cluster analysis of Prosopis strombulifera genotypes based on AFLP data generated with four primer pairs.

Fig. 3. Estimation of hypothetical cluster number (6) based on the inflection point (IP) of within-cluster sum of squares (WCSS) for diverse cluster arrangements
Discussion
AFLP markers were used to evaluate the degree of genetic diversity among samples in a natural population of P. strombulifera. The results show that specific combinations of selective primers made it possible to obtain a high number of polymorphic bands.
Genetic variability studies in many populations of Prosopis sp. have allowed, through efficient selection and multiplication tasks, to obtain outstanding fast-growing genotypes, with high biomass production and resistance to different types of stress. These improved genotypes judiciously combined with, for example, appropriate harvesting and water conservation techniques, drought hardening techniques in nursery stage, etc. and the correct choice of suitable sites, may allow forestation and reforestation tasks in various areas of the world, with greater chances of success (Golubov et al., 2001).
Knowing the degree of intra-population diversity in P. strombulifera studied by AFLP technique in this work, allows a better understanding of the great variability observed in the physiological responses of this species to salt treatments with different ion composition and osmotic potentials (Sosa et al., 2005; Llanes et al., 2005; Reginato, 2009; Llanes, 2010).
Our results show that individuals randomly collected from a native population can be defined using a hierarchical approach as belonging to five different genetic groups; otherwise, the use of a non-hierarchical approach points towards a population structure with at least six diverse groups. Despite differences between estimations of the number of putative genetic groups, the allocation of individual AFLP-profiles into larger and smaller clusters points towards a heterogeneous population structure for P. strombulifera, indicating a genetic differentiation among accessions. The variability found among them is evident both in the composition and the number and frequency of bands.
The polymorphism obtained in our study is comparable with variability index in populations of P. ferox (Burghardt et al., 2004) and P. ruscifolia (Burghardt & Palacios, 1998), and therefore justifies the conservation of this species as part of a gene bank approach. Some authors presented evidence to support the hypothesis that plants with predominant vegetative propagation, as is the case of P. strombulifera, have heterozygosis values higher than expected if the species is reproduced by sexual reproduction (Lacadena, 1970). In this sense, it is important to note that short inflorescences P. strombulifera are visited by many insects that may allow fertilization (Chiappa et al., 1997). Species whose seeds are dispersed by animals or the wind maintain high levels of variability within populations (Hamrick & Godt, 1990). Also, the high genetic diversity could be related to the geographical distribution. Species out crossing within a wide geographic range have higher levels of genetic diversity in relation to endemic species.
In conclusion, the results presented in this work in P. strombulifera native from a high-salinized area of central Argentina (Salinas El Bebedero, San Luis Province), add to the knowledge on the genetic diversity of this species of section Strombocarpa (Saidman et al., 1996). The observation of AFLP-profile differentiation among accessions may have many implications for conservation purposes, providing further support for genetic data collection and integration from this geographical region recognized as a valuable genetic source of Prosopis species.
In addition, maintenance of high levels of genetic diversity has adaptive significance for several species, and in this case, it may be absolutely necessary for P. strombulifera to face extreme environmental conditions such as lack of water, high temperatures and salt toxicity, among others (Reginato, 2009; Llanes, 2010).
Acknowledgements
This study was supported with funds from CONICET PIP 5628, PICTO-ANPCYT-UNRC 30093, ICGBE-TWAS Joint Biotechnology Programme, SECYT- Universidad Nacional de Río Cuarto and Ministerio de Ciencia y Tecnología de la Provincia de Córdoba (R 1210/2007) to Virginia Luna.
Bibliography
1. ALONSO-BLANCO, C., A. PEETERS, M. KOORNNEEF, C. LISTER, C. DEAN, N. VAN DEN BOSCH, J. POT & M. KUIPER. 1998. Development of an AFLP based linkage map gene-coding regions are found at z5-kb intervals; long of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 14: 259-271. [ Links ]
2. ANDERSON, D., J. DEL AGUILA & E. BERNARDON. 1970. Las formaciones vegetales en la provincia de San Luis. Revista Invest. Agropec., Ser. 2, 7:153-183. [ Links ]
3. BECKER, J., P. VOS, M. KUIPER, F. SALAMINI & M. HEUN. 1995. Combined mapping of AFLP and RFLP markers in barley. Mol. Gen. Genet. 249: 65- 73. [ Links ]
4. BENSCH, S. & M. AKESSON. 2005. Ten years of AFLP in ecology and evolution: why so few animals? Mol. Ecol. 14: 2899-2914. [ Links ]
5. BURGHARDT, A., S. ESPERT & R. BRAUNWILKE. 2004. Variabilidad genética en Prosopis ferox (Mimosaceae). Darwiniana 42: 31-36. [ Links ]
6. BURGHARDT, A. & R. PALACIOS. 1998. Variabilidad intraespecífica en Prosopis ruscifolia Griseb. (Leguminosae). Physis 55: 49-57. [ Links ]
7. BURKART, A. 1976. A monograph of the genus Prosopis (Leguminosae-Subfam. Mimosoideae). J. Arnold Arboret. 57: 219-249, 450-525. [ Links ]
8. CANTERO, J., A. CANTERO & J. CISNEROS. 1996. La vegetación de los paisajes hidrohalomórficos del centro de Argentina. Editorial Universidad Nacional de Río Cuarto, Río Cuarto. [ Links ]
9. CAROSIO, M., M. JUNQUERAS, A. ENDERSEN, M. FERNANDEZ BELMONTE, E. MARTINEZ CARRETERO, E. & A. DALMASSO. 2009. Flora de las Salinas del Bebedero. Sociedad de Biología de Cuyo, San Luis. Biocell 34: 1. [ Links ]
10. CHIAPPA, E., R. VILLASEÑOR, H. TORO & R. COVARRUVIAS. 1997. Reproductive tactics of Prosopis (Mimosaceae) and the ecological associations of its pollinizers in the northern desert of Chile. Multequina 6: 9-20. [ Links ]
11. DELLAPORTA, S., J. WOOD & J. HICKS. 1983. A plant DNA mini preparation: version II. Plant Mol. Biol. Rep. 1: 19-21. [ Links ]
12. EGAN, T. & I. UNGAR. 1998. The effects of different salts of sodium and potassium on the growth of Atriplex prostrate (Chenopodiaceae). J. Plant Nutr. 21: 2193-2205. [ Links ]
13. FU, Y., Y. FERDNÁNDEZ, A. PHAN, B. COULMAN & K. RICHARDS. 2004. AFLP variation in four blue grama seed sources. Crop Sci. 44:283-288. [ Links ]
14. GOLUBOV, J., M. MANDUJANO & L. EGUIARTE. 2001. The paradox of mesquites (Prosopis spp.): invading species or biodiversity enhancers?. Bol. Soc Bot. México 69: 23-30. [ Links ]
15. HAMRICK, J. & M. GODT. 1990. Allozyme diversity in plant species. In: BROWN, A., M. CLEGG, A. KAHLER & B. WEIR (eds.), Plant population genetics, breeding, and genetic resources, pp 43-63. Sinauer, Sunderland. [ Links ]
16. LACADENA, J. 1970. Genética Vegetal. Fundamentos de su Aplicación. AGESA, Madrid. [ Links ]
17. LARSON, S., T. JONES, Z. HU, C. MCCRACKEN & A. PALAZZO. 2000. Genetic diversity of bluebunch wheatgrass cultivars and a mulitiple-origin polycross. Crop Sci. 40: 1142-1147. [ Links ]
18. LLANES, A. 2010. Indicadores fisiológicos y moleculares de la tolerancia a salinidad en Prosopis strombulifera. Su correlación con los niveles endógenos de ABA. PhD Thesis, Universidad Nacional de Río Cuarto, Córdoba. [ Links ]
19. LLANES, A., H. REINOSO & V. LUNA. 2005. Germination and early growth of Prosopis strombulifera seedlings in different saline solutions. World J. Agric. Sci 1: 120-128. [ Links ]
20. LÓPEZ, M., J. MORENO & A. RAMOS-CORMENZANA. 2001. The effect of olive mill wastewaters variability on xanthan production. J. Appl. Microb. 90: 829-835. [ Links ]
21. MANIVANNAN, P., C. JALEEL, A. KISHOREKUMAR, B. SANKAR, R. SOMASUNDARAM & R. PANNEERSELVAM. 2008. Protection of Vigna unguiculata (L.) Walp. plants from salt stress by paclobutrazol. Colloids and surfaces B: Biointerfaces 61: 315-318. [ Links ]
22. MEUDT, H. & A. CLARKE. 2007. Almost forgotten or latest practice? AFLP applications, analyses and advances. Plant Sci. 12: 106-114. [ Links ]
23. PASIECZNIK, N., P. FELKER, P. HARRIS, L. HARSH, G. CRUZ, J. TEWARI, K. CADORET, & L. MALDONADO. 2001. The Prosopis juliflora- Prosopis pallida Complex: A monograph. HIDRA, Coventry, UK. [ Links ]
24. PEÑA ZUBIATE, C., D. ANDERSON, M. DEMMI, J. SAENZ & A. D´HIRIART. 1998. Carta de suelos y vegetación de la Provincia de San Luis. Secretaria de Agricultura, Ganadería, Pesca y alimentación. INTA, Estación Experimental Agropecuaria San Luis. [ Links ]
25. PHAN, A., Y. FU & S. SMITH. 2003. RAPD variations in selected and unselected blue grama populations. Crop Sci. 43:1852-1857. [ Links ]
26. REGINATO, M. 2009. Respuesta de la halófita Prosopis strombulifera a diferentes medios salinos. Modificaciones de los parámetros morfofisiológicos y su regulación hormonal. PhD Thesis. Universidad Nacional de Río Cuarto, Córdoba. [ Links ]
27. REINOSO, H., L. SOSA, L. RAMÍREZ & V. LUNA. 2004. Salt-induced changes in the vegetative anatomy of Prosopis strombulifera (Leguminosae). Can. J. Bot. 82: 618-628. [ Links ]
28. REINOSO, H., L. SOSA, M. REGINATO & V. LUNA. 2005. Histological alterations induced by sodium sulfate in the vegetative anatomy of Prosopis strombulifera (Lam.) Benth. World J. Agric. Sci. 1: 109-119. [ Links ]
29. SAIDMAN, B., J. VILARDI, M. POCOVI & N. ACRECHE. 1996. Genetic divergence among species of the section Strombocarpa, genus Prosopis (Leguminosae). J. Genet. 75: 139-149. [ Links ]
30. SOSA, L. 2005. Adaptaciones fisiológicas de Prosopis strombulifera a condiciones de salinidad por cloruros y sulfatos. PhD Thesis, Universidad Nacional de Río Cuarto, Córdoba. [ Links ]
31. SOSA, L., A. LLANES, M. REGINATO, H. REINOSO & V. LUNA. 2005. Osmotic and Specific Ion Effects on the Germination of Prosopis strombulifera (Lam.) Benth. Ann. Bot. 96: 261-297. [ Links ]
32. VOS, P., R. HOGERS, M. BLEEKER, M. REIJANS, T. VAN DE LEE, M. HORNES, A. FRIJTERS, J. POT, J. PELEMAN, M. KUIPER & M. ZABEAU. 1995. AFLP: a new technique for DNA fingerprinting. Nucl. Acids Res. 23: 4407-4414. [ Links ]
33. WANG, Y., C. THOMAS & R. DEAN. 1997. A genetic map of melon (Cucumis melo L.) based on amplified fragment length polymorphism (AFLP) markers. Theor. Appl. Genet. 95: 791-798. [ Links ]
34. WAUGH, R., N. BONAR, E. BAIRD, B. THOMAS, A. GRANER, P. HAYES & W. POWELL. 1997. Homology of AFLP products in three mapping populations of barley. Mol. Gen. Genet. 255: 311- 321. [ Links ]
35. WITTEN, I. & E. FRANK. 2005. Data Mining: Practical Machine Learning Tools and Techniques. Morgan Kaufmann, San Francisco. [ Links ]
Recibido el 10 de Febrero de 2011,
aceptado el 11 de Agosto de 2011.














