Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
Boletín de la Sociedad Argentina de Botánica
versión On-line ISSN 1851-2372
Bol. Soc. Argent. Bot. vol.49 no.4 Córdoba dic. 2014
GENÉTICA Y EVOLUCIÓN
Morphological and molecular characterization of common bean landraces cultivated in the semi-arid Mexican high plateau
Homar Rene Gill Langarica1*, Rigoberto Rosales Serna2, Sanjuana Hernandez Delgado1 and Netzahualcoyotl Mayek Perez1
1 Instituto Politécnico Nacional-Centro de Biotecnología Genómica, Boulevard del Maestro Esquina Elias Piña. CP. 88710, Reynosa Tamaulipas, Mexico. *E-mail: hgill@ipn.mx
2 Campo Experimental Valle del Guadiana-Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias (INIFAP). Carretera Durango-El Mezquital km 4.5, Durango, Dgo. México. CP. 34170.
Summary
The objective of the current research was to characterize the common bean in agricultural fields planted with common bean landraces from the semi-arid Mexican high plateau using both morphological and amplified fragment length polymorphism (AFLPs) data. The morphological traits were discriminating and exhibited the clustering of 150 accessions based on the geographic origin and seed coat color. AFLP primer combinations exhibited a polymorphic range between 0.292 (E-AGG + M-ACT) and 0.375 (E-ACA + M-AGA). The frequency and distribution of the polymorphic fragments allowed the detection of a larger number of rare fragments in accessions 121 and 111 (Flor de Mayo and black common beans, respectively). The analysis of genetic relationships, analysis of molecular variance (AMOVA), and Powell's diversity index confirmed a broad genetic basis for the germplasm of the common bean from the semi-arid Mexican high plateau. The clustering and principal coordinate analyses demonstrated a strong genetic relationship among the collected common bean landraces based on the similarity in the variety name, origin, and seed coat color, demonstrating the influence of different cultivation practices in the two regions and the adaptation of P. vulgaris to the agroclimatic conditions of the semi-arid Mexican high plateau.
Key words: AFLP; Genetic variation; Landraces; Phaseolus vulgaris L.
Resumen
Caracterización morfológica y molecular de las variedades locales de frijol común cultivado en el altiplano mexicano semi-árido. El objetivo de este trabajo fue caracterizar el frijol común de parcelas agrícolas cultivadas con variedades criollas de frijol común de la región semiárida del altiplano mexicano utilizando datos morfológicos y de Polimorfismos en la Longitud de los Fragmentos Amplificados (AFLPs). Las características morfológicas fueron discriminantes y mostraron la agrupación de las 150 accesiones en función del origen geográfico y el color de la testa de la semilla. La combinación de iniciadores AFLP (amplified fragment length polymorphism) mostraron un rango polimórfico entre 0,292 (EM-ACT AGG +) y 0,375 (M + E-ACA-AGA). La frecuencia y distribución de los fragmentos polimórficos permitió la detección de un mayor número de fragmentos raros en las accesiones 121 y 111 (Flor de Mayo y frijol común Negro, respectivamente). El análisis de las relaciones genéticas, varianza molecular (AMOVA), y el índice de diversidad de Powell confirmaron amplia base genética del germoplasma de frijol común del altiplano semi-árido mexicano. El análisis de coordenadas principales y de agrupación demostraron fuerte relación genética entre las variedades colectadas con base a la similitud en el nombre de la variedad, el origen y el color de la testa de la semilla, lo que demuestra la influencia de las diferentes prácticas de cultivo en las dos regiones y la adaptación de P. vulgaris a las condiciones agroclimáticas de la región semiárida del altiplano mexicano.
Palabras clave: AFLP; Criollos; Phaseolus vulgaris L.; Variación genética.
Introduction
The common bean (Phaseolus vulgaris L.) is an ancient crop that originated on the American continent. The origin of the common bean is corroborated by the cultivation methods implemented by a wide variety of settlers and farmers, the uses of the crop, and the range of environments to which the common bean has adapted (Broughton et al., 2003). The common bean has been growing in Mexico since the pre- Hispanic period, and Mexico is known as the main center of domestication for the common bean when regarding the Mesoamerican genetic pool of this species, which exhibits a high genetic diversity (Gepts& Debouck, 1991), and as a center of origin for the common bean (Bitocchi et al., 2011).
The genetic diversity of the common bean in Mexico is safeguarded at the National Institute of Forestry, Agricultural, and Livestock Research (INIFAP, Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias). INIFAP has used this diversity to generate improved varieties using cultivated strains, landraces, and wild samples as progenitors (Acosta et al., 1999). This practice has led to the development of new and improved commercial classes of common beans, such as the Azufrado and Peruano varieties (Rosales-Serna et al., 2004).
In rural areas of Mexico, the common bean landraces are still cultivated and are valued only in local markets. These landrace germplasms of common bean are classified by the farmer based on the seed color and flavor and are identified using specific names. It is common for commercial common bean varieties grown in Mexico to be identified by the specific names of their landrace ancestors because the systematic identification of these varieties using specific names is an important aspect of the management of these commercial varieties within local agricultural systems. It is not clear if these varieties maintain their genetic identity, if the genetic variation is influenced by the distribution patterns, or if the names associated with specific phenotypes can be used to evaluate the genetic diversity of the common beans available in a particular region.
Until now, all research conducted in the semi-arid Mexican high plateau based on common bean genetic resources has been based on morphological and phenological characterization (Rosales-Serna et al., 2003), and the phenotypic evaluation has demonstrated high value. Molecular techniques, such as amplified fragment length polymorphism (AFLP) marker technology (Vos et al., 1995), have been effective tools for studying the genetic relationships among common bean germplasms (Rosales-Serna et al., 2005; Kumar et al., 2008; Perseguini et al., 2011) and are being used extensively for studies on plant genetic diversity (Tatikonda et al., 2009; Pecina-Quintero et al., 2011; Boczkowska et al., 2012). The objective of the current research was to investigate the status of common bean landrace genetic resources grown in agricultural parcels of the semi-arid Mexican high plateau based on morphological and AFLP data. This research was also motivated by the assumption that genetic diversity in common bean is prone to change; therefore, there is a possibility for a decrease or increase in the genetic diversity of common bean because of the constant movement of materials throughout Mexico.
Materials and Methods
Study area and collections
The study area consisted of three states located in the semi-arid Mexican high plateau (Chihuahua, Durango, and Zacatecas) in the north of Mexico. For comparative purposes, landrace samples from the states of Hidalgo, Puebla, and Guanajuato were included. In 2006, 10 individual plants were collected from each of 12 agricultural parcels sown with common bean landraces in the states of Durango (4), Chihuahua (4), Zacatecas (4), Hidalgo (1), Puebla (1), and Guanajuato (1) (Table 1).
Table 1. Relationship of common bean (Phaseolus vulgaris L.) landraces analyzed in the semi-arid Mexican high plateau.

† External evaluation group.
Morphological analysis
In 2007, agronomic characterization was carried out following specific design protocols that were adopted from previously used field trial outlines; the data scoring protocol was adapted from the standard evaluation protocol of the Mexican National Seed Inspection and Certification Service (SNICS, Sistema Nacional de Inspección y Certificación de Semillas, 2001) for the common bean. Ten plants from each accession were planted in rows (5 m long) with three replications in the Campo Experimental Valle del Guadiana in Durango state, which belongs
to the Mexican National Institute of Forestry, Agriculture, and Livestock Research (INIFAP).
In total, 51 qualitative and quantitative traits of the leaves, pod, seed, and architecture of each plant were measured, as was the reaction of the common bean plants to disease (Table 2). The traits evaluated in each accession were classified into the following five categories: 1) phenological traits, 2) plant architecture and yield components (International Board for Plant Genetic Resources, 1982), 3) color of the recently harvested dry sedes (van Schoonhoven& Pastor-Corrales, 1987), 4) seed quality (van Schoonhoven& Pastor- Corrales, 1987), and 5) reaction to diseases that are common and cause destructive damage in the test site. These diseases included anthracnose disease [Colletotrichum lindemuthianum (Sacc. and Magn.) Scribner], halo blight [Pseudomonas syringae pv. phaseolicola (Burkholder)], common bacterial blight [Xanthomonas campestris pv. phaseoli (Smith) Dye], bean common mosaic virus, root rot (Rhizoctonia solani Kühn), and angular leaf spot [Phaeoisariopsis Griseola (Sacc.) Ferr.]. The damage analysis was performed during the reproductive phase (flowering of each accession) and in accordance with the standard grades provided by the International Center for Tropical Agriculture (CIAT, Centro Internacional de Agricultura Tropical) (van Schoonhoven& Pastor-Corrales, 1987), which includes nine destructive grades (1-9) in which 1= no symptoms and 9 = greater than 75% of the plant exhibits disease symptoms. The values from 1 to 3, 4-6, and 7-9 are classified as indicating resistance reactions, intermediate reactions, and susceptibility to the pathogen, respectively.
Germplasm was classified by geographic origin (state) and commercial-seed class (color) (Table 1). In Mexico, there are approximately 16 commercial classes of common beans according to the seed color: red mottled (Cacahuate); purple (Morado); black (Negro); marbling (Jaspeado); yellow (Amarillo); cream (Bayo); brown-striped (Ojo de Cabra); cream mottled (Pinto); white (Blanco); white mottled (Vaquita); light purple (Manzano); brown (Café); pink (Flor de Mayo); red (Rojo); gray (Gris); pink-striped (Flor de Junio) (Bellón et al., 2009; Rosales- Serna et al., 2004; SNICS, 2001; Voysest, 2000; Cárdenas, 1984).
With the existence of a physical support in the field experiment to allow the plants to climb and conform to a type IV (climbing) habit of growth, growth habit types were classified differently from those reported by Debouck& Hidalgo (1982): Ia, bushy determined, stem and branches erect and strong; Ib, bushy determined, weak stem and branches; IIa, erect indeterminate or undeveloped short guides; IIb, erect indeterminate guides, medium to long, non-climbing; IIIa, indeterminate prostrate, non-climbing short guides; IIIb, indeterminate prostrate long guides, climbers; IVa, climber indeterminate, pods distributed throughout the plant; IVb, climber indeterminate, pods concentrated at the top of the plant. Quantitative traits were measured as follows: The number of seeds per pod was scored as low (≤ 4), medium (5-6), or high (≥ 7); the flowering period was very early (<40 days), early (40-45 days), mid (45-50 days), delayed (50-60 days), or very late (> 60 days); the 100-seed weight was small (<25 g), medium (25-40 g), or large (> 40 g); plant height was low (<30 cm), medium (30-50 cm), or high (> 50 cm); pod length was very short (<4 cm), short (4-7 cm), medium (7-10 cm), long (10-13 cm), or very long (> 13 cm); pod width was small (<6 mm), medium (6-10 mm), or large (> 19 mm); and leaflet tip size was small (<5 cm), medium (5-9 cm), or large (> 9 cm). Intervals were used for quantitative traits to adjust the morphological characterization guidelines of the International Union for the Protection of New Varieties of Plants (UPOV) for application to P. vulgaris.
AFLP analysis
Plant material
All common bean landraces were selected to investigate variation within and between farmer´s materials (Table 1). Five seedlings were obtained from each of the 10 individual plants from each of the 15 agricultural parcels and bulked together. Their DNA was extracted, resulting in 150 DNA samples in total.
Molecular procedures
To obtain young tissue from the accessions, the seeds were germinated in trays using a sterile substrate. Two weeks after emergence, the samples were collected. The total genomic DNA was isolated using the method described by Dellaporta et al. (1983). The analysis of the polymorphisms in the length of the amplified fragments was performed as described by Vos et al. (1995). First, the genomic DNA was digested using 5 U EcoRI and MseI (Roche®) at 37 °C for 3 hours. Next, the digested DNA samples were incubated at 70 °C for 15 minutes to deactivate the restriction enzyme. Finally, the adapters [5 pmol EcoRI and 50 pmol MseI] were added to the digested DNA fragments along with the ligation buffer (1x T4 DNA ligase) and 1 U T4 DNA ligase and incubated at 37 °C overnight. The AFLP loci were amplified using four primer pair combinations (E-AGG + M-ACT, E-ACT + M-CTA, E-ACA + M-AGA, and E-ACC + M-AGA). The selective pre-amplification was performed using complementary primers for the EcoRI and MseI adapters, using two selective nucleotide bases (adenine and cytosine). The PCR was performed in a Px2 thermocycler (Thermo Electron Corporation, Milford, MA, USA). The selective amplification was performed using EcoRI and MseI primers labeled with fluorescent dye (IRD700 and IRD800) and three selective nucleotides. Each of the PCR products was electrophoresed on denaturing polyacrylamide gels (6 %). The PCR products were separated in a sequencing system (Li-COR IR2; LI-COR, Inc. Lincoln, NE, USA) equipped with an infrared laser with the capacity to read wavelengths between 700 and 800 nm. The profile of the obtained fragments was analyzed using Cross Checker V.2.9 software (Buntjer, 1999).
Statistical analysis
Morphological data
To visualize the existing relationships between the accessions of the common bean from the semi-arid Mexican high plateau and the external sample groups, the morphoagronomic data were used to create a matrix of quantitative and standardized qualitative data. This matrix was used to build a dendrogram based on the similarity coefficient for multistate variables described by Rogers & Tanimoto (1960) and the unweighted pair group method with arithmetic mean algorithm. To corroborate the interrelationships between the common bean samples, a tridimensional graphical representation was built using principal coordinate analysis (PCooA) with the similarity coefficient of Rogers & Tanimoto (1960). The analyses were performed using NTSYSpc statistical software V2.2 (Rohlf, 2009).
AFLP data
To determine the most informative combination of AFLP primers, parameters such as the polymorphic information content (PIC), marker index (MI), and resolution power (RP) were analyzed (Laurentin& Karlovsky, 2007). The PIC value for each AFLP primer pair was calculated as described by Roldan et al. (2000); PIC = 2ƒi (1 - ƒi), where PIC is the polymorphic information content of marker i, ƒi is the frequency of markers present, and 1 - ƒi is the frequency of absent markers. The MI was calculated using the formula from Varshney et al. (2007): MI = PIC x EMR, where EMR is the effective multiplex ratio (E), defined as the total number of loci products/ number of fragments per primer (n) multiplied by the fraction of polymorphic loci/number of fragments (β) ( E = nβ). The RP of each primer was calculated according to Prevost and Wilkinson (Prevost& Wilkinson, 1999) as follows: RP = ΣIb, where Ib represents the informativity of the fragment. The Ib term contains values from 0 to 1 and is determined by the formula Ib = 1-[2 x |0.5 - p|], where p is the proportion of the 150 samples contained the fragment.
Diversity index
The germplasm genetic diversity index was calculated using the formula described by Powell et al. (1996); DI = 1 - ΣP2i, where Pi is the frequency of the in allele, and each individual allele is considered a unique amplified fragment.
Intra and Inter-cultivar Variation and Genetic Identification
The common bean landraces were selected according to the seed coat color and agricultural parcel origin for the analysis of variation among and between the farmer samples (Table 1). A binary matrix was used to calculate the molecular variation analysis (AMOVA) (Huff et al., 1993) based on the following hierarchical ordering: groups (Chihuahua, Durango, Zacatecas, Hidalgo, Guanajuato, and Puebla) and populations within groups (common bean types within states). The number of AMOVA significance test permutations was 1,023 (Guo& Thompson, 1992). AMOVA was performed using the Arlequin statistical program V.3.5 (Excoffier& Lischer, 2010). To determine existing genetic relationships between the 150 common bean samples analyzed, a binary matrix was created (where 1 denotes the presence and 0 the absence of a locus) for each pair of AFLP primers. A dendrogram was calculated using the Jaccard similarity coefficient (Nei& Li, 1979) and the weighted neighbor-joining algorithm and jackknife method for corroboration. This resampling method was applied to determine the robustness of the dendrogram. A resampling of 10,000 repetitions was performed based on the original data from the 150 common bean accessions. To corroborate the interrelations, a tridimensional graphical representation was generated, and PCooA was performed using the Jaccard similarity coefficient (Nei& Li, 1979). The clustering analysis was performed using the DARWIN V5 statistical software (Perrier& Jacquemoud, 2006), and the NTSYSpc statistical program V.2.2 (Rohlf, 2009) was used for the PCooA.
Results
Morphological traits
The UPGMA clustering analysis of the common bean landraces from the semi-arid Mexican high plateau demonstrated two main germplasm groups (Fig. 1). Group 1 consisted of the Flor de Mayo (external group) common bean variety from the agricultural parcels in Hidalgo and Guanajuato states, Flor de Mayo Media Oreja from the parcels located in Chihuahua, Flor de Junio common bean variety from the parcels in Zacatecas, and cream-colored common beans with brown speckles (Pinto and Ojo de Cabra common bean landraces) collected in the agricultural parcels in Chihuahua. The second group consisted of black common beans from the parcels in Durango and Zacatecas, Bayo variety common beans from Zacatecas, Canario variety common beans from the two parcels in Durango, and Palacio variety common beans from Puebla state.

Fig. 1. Graphical representation of genetic similarities based on the Roger and Tanimoto similarity coefficient for the germplasm of the common bean in from the semi-arid Mexican high plateau, based on 51 morpho-agronomic traits. The numbers 1 and 2 refer to the clusters discussed in the text.
The germplasm was represented by a scatter plot using the results of the PCooA. The following three principal groups were observed (Fig. 2):

Fig. 2. Scatter plot representation of common bean germplasm from the semi-arid Mexican high plateau analyzed based on PCooA and 51 morpho-agronomic traits. The blue shape contains material from Durango state; red, from Chihuahua state; green, from Zacatecas state; pink, from Hidalgo state; melon, from Guanajuato state; and pistachio-green plus cross, from Puebla state. The numbers 1, 2 and 3 refer to the clusters discussed in the text.
Group 1 included pink and cream common beans from parcels located in Chihuahua [Pinto (two parcels), Ojo de Cabra, and Flor de Mayo Media Oreja] and Durango (Flor de Junio), and Flor de Mayo from agricultural parcels in Hidalgo and Guanajuato. Group 2 consisted of only black common beans from Durango and Zacatecas (two parcels). Group 3 included Canario common beans from Durango (two parcels), Bayo common beans from Zacatecas, and Palacio common bean landraces from Puebla.
AFLP analysis
In this study, 4 different AFLP primer pairs were used, and each AFLP primer pair generated information on the 120 common bean landrace accessions collected from the different agricultural parcels located in the semi-arid Mexican high plateau and the 30 landrace accessions collected from parcels located in the states of central Mexico. The number of products amplified per combination varied from 61 to 130. In total, the 4 combinations of selected primers amplified 406 products, of which 382 (94 %) were polymorphic.
The 382 polymorphic fragments were classified as unique and rare fragments. Unique fragments are specific to an accession by AFLP combination, and five unique fragments were observed. The accessions with identification numbers 55 and 151 for Ojo de Cabra and Palacio from Chihuahua and Puebla, respectively, demonstrated two unique alleles in different AFLP combinations (E-ACA + M-GTA and E-ACC + M-AGA). The number of rare fragments was determined and defined as the fragments present in only 10 % of accessions via AFLP combination. In total, 50 fragments were observed in 150 common bean landrace samples, with an average of 12.5 per AFLP combination. A larger number of rare fragments (22) were observed in the E-AGG + M-ACT combination, whereas the E-ACA + M-AGA combination demonstrated only fragments. Of the 150 common bean landrace accessions analyzed, the Flor de Mayo accessions 121 and 128 from Hidalgo, and the black common bean accession 111 from Zacatecas had the largest number of rare fragments.
To identify the most informative AFLP primer combinations, the PIC was determined, exhibiting a range per combination between 0.292 (E-AGG + M-ACT) and 0.375 (E-ACA + M-AGA), with an average of 0.336 per AFLP combination. EMR ranged from 77 to 116, with an average of 95.3 per AFLP combination. The E-ACC + M-AGA and E-ACT + M-CTA combinations demonstrated the highest and lowest values, respectively. MI ranged from 28.56 to 36.37 with a median of 31.81 per combination. The E-ACC + M-AGA combination demonstrated the highest value and E-ACT + M-CTA the lowest value. To evaluate the discriminatory power between AFLP combinations, the RP was determined, which ranged from 41.3 to 57.2 with an average of 49.8. The E-ACA + M-AGA combination demonstrated the highest value and E-AGG + M-ACT the lowest value. The Powell genetic diversity index varied from 21% to 29% per combination with an average of 24%. The E-ACA + M-AGA combination demonstrated the highest value, and the E-ACC + M-AGA and E-AGG + M-ACT the lowest value.
Intra- and Inter-cultivar Variation and Genetic Identification
The AMOVA of the AFLP data demonstrated that 71.2% of the genetic variation occurred within populations (accessions), 17% occurred between populations within regions, and 11.7% between regions. Therefore, the highest proportion of genetic variation was demonstrated within populations and not between regions. The dendrogram (Fig. 3) shows the formation of two principal accession groups. Although there was a tendency to form groups by geographic origin or bean type (seed coat color), the dendrogram consisted of groups of different regions, which suggests a broad genetic base for the common bean in the semi-arid Mexican high plateau.
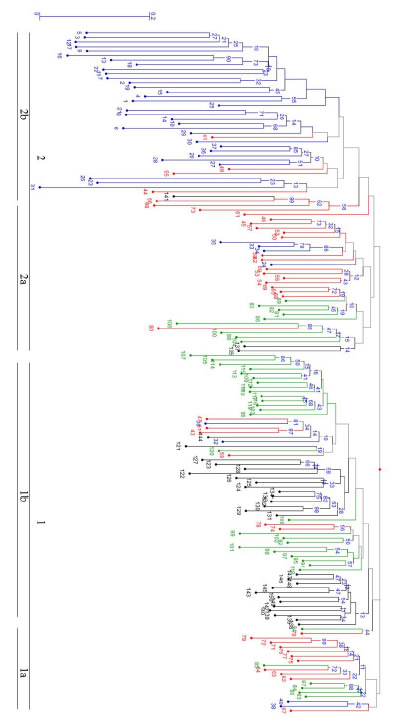
Fig. 3. Graphical representation of genetic similarities of common bean germplasm from the semi-arid Mexican high plateau based on the Jacard similarity coefficient and AFLP markers. The values in the nodes indicate bootstrap values after 10,000 repetitions. The numbers 1, 2 and 1a, 1b and 2a, 2b refer to the clusters and subclusters discussed in the text.
Group 1 was the most heterogeneous and was subdivided into two subgroups (1a and 1b). Subgroup 1a was made up of 17 accessions: Pinto variety common bean landraces (accessions 62, 63, 64, and 67) from Lázaro Cárdenas; Flor de Mayo Media Oreja common beans (accessions 71, 72, 75, 76, 77, and 79) from Benito Juarez, Chihuahua; and Bayo common beans (accessions 84, 85, 87, and 90) from Rio Grande, Zacatecas. Subgroup 1b had the highest number of accessions (62 in total). This subgroup combined common beans from the external collections from central Mexico, such as Flor de Mayo and Palacio (from parcels in Guanajuato, Puebla, and Hidalgo) and common beans from Zacatecas (4 locations; Bayo, black, and Flor de Junio common beans). This subgroup also included accessions from Lázaro Cárdenas (Pinto common bean landrace accessions 43 and 47 and Ojo de Cabra common bean accessions 57 and 59); Namiquipa, Chihuahua (Flor de Mayo Media Oreja common bean accessions 74 and 78); and Poanas la Joya, Durango (Flor de Junio common bean accessions 32 and 39).
Group 2 was more homogenous and comprised two subgroups (subgroup 2a and 2b). Subgroup 2a was formed by 30 accessions, mostly comprising Pinto variety common beans from the Lázaro Cárdenas community in Chihuahua; Bayo and Negro variety common beans from Rio Grande and González Ortega in Zacatecas, respectively; Flor de Junio and Negro variety common beans from Poanas la Joya in Durango; and two accessions (135 and 137) from an external group of samples of Flor de Mayo variety common beans from San Luis de la Paz in Guanajuato. Subgroup 2b contained the largest number of samples (accession 41), mostly Canario, black, and Flor de Junio common bean landraces collected from agricultural parcels in Durango (Ricardo Flores Magon, Antonio Amaro, and Poanas la Joya), Pinto and Flor de Mayo Media Oreja samples from Lázaro Cárdenas and Namiquipa in Chihuahua, and accession no. 141 Palacio bean variety from Puebla. The clustering analysis results were confirmed using PCooA analysis as shown in Figure 4, which demonstrates that 71% of the genotypic variation was explained by the spatial separation of the common bean samples.
Discussion
For most characters, great diversity was found among the accessions. This diversity is largely continuous, with a clear separation in the three-dimensional and dendrogram representation (Figs. 1 and 2). The growth habit trait showed the greatest number of classes in the architecture of the plant and was predominantly indeterminate, with non-climbing shoots. These results agree with those obtained in similar studies by Rosales-Serna et al. (2001) and Garcia et al. (1997). The primary pod color was yellow in all of the accessions, but the secondary pod was green, red, or colorless. The predominant seed types were Flor de Mayo, Pinto and black, with differing intensities (Table 2). These are the preferred colors of common beans by farmers and consumers in Mexico (Bellon et al., 2009). Most black bean accessions came from the center region of Mexico. This fact is influenced by the preference for black common beans in this region. In a recent study, Andean seed colors were distributed as 23% cream, 17% black, 11% yellow, and 49% other (Blair et al., 2009). As expected, all accessions with purple flowers had black seeds, in accordance with Bassett (1995), who reported that purple and pink flowers confer black seed coats. Many farmers select seeds for sowing based on the seed size and color (Papa& Gepts, 2003).
Table 2. Morphological qualitative traits evaluated in the sample of semi-arid Mexican high plateau common bean (Phaseolus vulgaris L.) landraces.

This study adds new insights into the picture of diversity recently drawn for the semi-arid Mexican high plateau common bean landraces (Beebe et al., 2000). First, the sample of common bean landraces presented significant diversity in its morphology, with very diverse seed colors, architecture, phenology, flowers and leaves. However, not all morphologies reported by IBPGR (1982) for the species had a wide variety of classes in this study; for example, the growth habit was only present in five of eight possible classes in this sample. These results are in agreement with a previous study performed in the same region with landrace samples (Rosales-Serna et al., 2001; Beebe et al., 2000).
Second, interesting traits were identified related to the nomenclature for common bean landraces in the semi-arid Mexican high plateau. Among the accessions of common bean landraces with shared common names in the present study, duplicates were only observed in outside reference accessions, for example, accessions 143 and 148 and accessions 141, 142, and 145 of Palacio common beans of Puebla (Fig. 1 and Table 1). Most of the accessions within the same varietal class (as indicated by a shared name) presented clear morphological, agronomic, and/or genetic differences, for example, in flower color (wing and standard), seed color, determinacy, and disease resistance. Vargas-Vazquez et al. (2006) also evaluated the morphological diversity among accessions in a common bean gene bank from INIFAP and observed that within the same varietal classes, there were no duplicates. Despite the fact that some diversification may have occurred within the same landrace type over a long period of cultivation, there is also the aspect of uncertainty of landrace names chosen by small farmers.
Third, the morphological diversity of the semi-arid Mexican high plateau group was either equal to or slightly higher than the diversity in outgroups materials, although the second was less numerous. With regard to agronomic traits, both groups in our sample of landraces also showed clear differences in their susceptibility and resistance to important common bean diseases. For example, both groups presented a low proportion of accessions susceptible to anthracnose, halo blight, root rot, and bean common mosaic virus resistance evaluated in field conditions. For angular leaf spot and common bacterial blight resistance, the semi-arid Mexican high plateau group (common bean Canario, Flor de Junio and Flor de Mayo types) included fewer resistance accessions than the out-group's materials (common beans Flor de Mayo and Palacio types), but both groups included greater amounts of materials with intermediate values of resistance to both pathogens.
The above observations indicate that the landrace samples in the northern region of the country were introduced from the central Mexican states and adapted to the environmental conditions of the semi-arid high plateau, which emphasizes the domestication of the Mexican common bean and the distribution of the species by the farmers and breeders. Similarly, as shown by Lopez et al. (2005), landraces and pre-improved forms of P. vulgaris demonstrate a lower range of environmental adaptation than the domesticated varieties managed by farmers and breeders, which has modified important traits of the plant, such as biological cycle, reaction to diseases, and plant structure. These adaptations have led to the development of a greater diversity in the environments and local consumer preferences, allowing the consumption of Pinto, Bayo, Flor de Mayo, and Flor de Junio commercial varieties in the semi-arid Mexican high plateau and, to a lesser extent, the black and Ojo de Cabra varieties (Acosta et al., 2002).
AFLP analysis allows for improved estimates of the genetic relationships between closely related individuals. In this study, the AFLP analysis detected high levels of polymorphism (94%) between 150 samples of common beans landraces. The analysis of the polymorphic fragment frequency and distribution demonstrated five specific fragments present in three accessions and 50 rare fragments present in up to 10% of the accessions. Genetic improvement programs for the common bean in Mexico often use landrace germplasm to obtain better results by contributing agronomically important and productive traits to existing commercial cultivars (Acosta et al., 1996; Acosta et al., 1999). Thus, this information may be useful for genetic improvement programs for common beans because it would aid the selection of materials that would increase variability or take advantage of heterosis (Acosta et al., 1999). A potential use for the analysis of the intra-population genetic variation of common beans using AFLPs is that the AFLP combinations demonstrating a higher number of rare and unique fragments (E-AGG + M-ACT and E-ACC + M-AGA) could be used to increase differences between accessions from the same populations. The highest number of rare and unique fragments were exhibited by the Pinto common beans (accession 55, Ojo de Cabra common bean landrace), Flor de Mayo (accessions 121 and 128), and black common beans (accession 111). The presence of these polymorphisms (unique and rare fragments) in one accession in particular would allow the development of specific sequences for the development of markers linked to quantitative trait loci of agronomic importance in P. vulgaris. For example, accession 55 (Ojo de Cabra common bean type) is highly resistant to anthracnose, halo blight, root rot, and bean common mosaic virus but is moderately tolerant to angular leaf spot and common blight, and it has a determinate growth habit and short production cycle (98 days to physiological maturity).
The PIC is used widely in genetic diversity studies. In the case of dominant markers (biallelic), such as AFLPs, the maximum expected value for the PIC is 0.50. In this study, the AFLP combinations demonstrated an average PIC value of 0.25. Based on the total PIC value (0.32), the use of the E-ACA + M-AGA combination is recommended for P. vulgaris landrace germplasm analysis because it differs from that reported by Rosales-Serna et al., (2005). The obtained MI value corroborated the usefulness of the E-ACA + M-AGA AFLP combination and demonstrated that the E-ACC + M-AGA combination might be of use in common bean germplasm analysis because these combinations demonstrated MIs of 33.38 and 36.37, respectively. Whereas the PIC has been used more widely for studying organisms of economic interest (Varshney et al., 2007; Gupta et al., 2008). Prevost& Wilkinson (1999) implemented the concept RP for evaluating the discriminatory effect of AFLP combinations. Therefore, the E-ACT + M-CTA and E-ACA + M-AGA combinations with high RP values (56.7 and 57.2, respectively) are more informative for differentiating related accessions (Prevost& Wilkinson, 1999).
Compared with the observed genetic diversity in other regions of Mexico (Rosales-Serna et al., 2005), the 150 landrace samples of common bean collected in the parcels from the semi-arid Mexican high plateau demonstrated medium to high levels of genetic diversity. These data confirm the presence of high genetic diversity in the landraces common bean germplasm in semi-arid Mexican high plateau region (Gepts& Debouck, 1991). Farmers from the semi-arid Mexican high plateau often grow different common bean types, which include the three or four P. vulgaris races (e.g., the Durango, Jalisco, Mesoamerican, and New Granada races) found in the region. These small-scale growers use plant seeds of two or more common bean landraces that have been carefully selected, sometimes mixing different seeds for cultivation (Delgado- Salinas et al., 2006). The type of cultivation practiced in the area is used mainly to mitigate the adverse effects of drought in the area, to increase harvest security (Beebe et al., 2000), and to improve or ensure the simultaneous adaptability of different pre-improved and improved materials to the semi-arid high plateau agro-ecosystem for sustainable common bean production (Bellon et al., 2009). These practices also improve disease management, especially when different levels of resistance are present in different cultivated common bean varieties (Cárdenas et al., 1996).
Whereas the common bean is predominantly an autogamous species, none of the accessions analyzed using AFLPs were identical (Fig. 3). The causes of the intra-variety variation in the landrace cultivated by the farmers possibly include a cross between common bean types from the same breed or common bean variety; a cross between different common bean breeds, which generated the existing genetic diversity in the semi-arid Mexican high plateau region; or spontaneous mutations (Ko et al., 1994). The genetic relationship between the common bean type grown by farmers of the different localities and states provides new information on the practices of variety identification by the farmers. A number of common bean types with the same name were grouped genetically, including those collected in different localities. Therefore, the traditional names of the common bean landrace types in the semi-arid Mexican high plateau might be a good indicator of the state of the genetic diversity in the landrace common bean in the northern region of the country. This hypothesis is consistent with a report by Appa Rao et al. (2002), which demonstrated that the names of varieties offer clues to the genetic diversity of rices in specific regions.
The results of the AFLP analysis demonstrated the genetic similarity between the common bean landraces with names identical to those in other regions. Farmers in different regions exchange materials (Cárdenas, 2000). The genetic similarity of common bean landraces with similar names demonstrates the migration effects of germplasm, and the genetic behavior may be because of similar cultivation practices used by the farmers in the different regions analyzed. The main differences observed were in the planting systems, cultural practices (such as crop density), level of variety mixtures, and mixed cultivation techniques (Cárdenas, 2000). These differences in cultural practices have apparently influenced the development of the common bean landrace germplasm, even those germplasms coming from the same region that change their genetic profile after growth in separate areas. This observation is consistent with reports by Teshome et al. (1999), in which crop management practices were shown to change the morphological and genetic identity of sorghum. The clustering and PCooA analyses (Figs. 3 and 4) demonstrated the formation of two principle accession groups, including external evaluation materials. These data suggest an intra-cultivar genetic base for the common bean germplasm in the semi-arid high plateau of northern Mexico. This germplasm base contained materials that may have few, and likely unique, genetic changes. These changes may be the result of the evolution of P. vulgaris in various environments or via the adaptation by farmers and breeders to the environmental conditions of the northern Mexican dry lands (Blair et al., 2011).
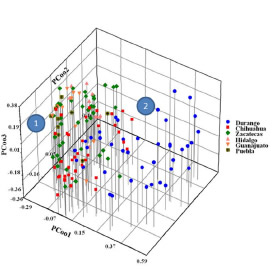
Fig. 4. Scatter plot representation common bean germplasm from the semi-arid Mexican high plateau analyzed based on PCooA and AFLP markers. The blue shape contains material from Durango state; red, from Chihuahua state; green, from Zacatecas state; pink, from Hidalgo state; melon, from Guanajuato state; and pistachio-green plus cross, from Puebla state. The numbers 1, 2 refer to the clusters discussed in the text.
By integrating the morphological, agronomic, and molecular characterization into the diversity assessment of a Mexican germplasm collection of common beans from the semi-arid Mexican high plateau, we were able to improve our understanding of the organization of this diversity in this region of northern Mexico. The integration of these different types of data into this assessment also allowed for the identification of important differences among the two genetic groups analyzed, such as for agronomic and genetic traits. Our results emphasize the importance of such an integrated approach in the diversity assessment for the conservation of resources, as a clue to promote the use of genetic resources of farmers, such as landrace germplasm collections. Finally, it is important to note that the two types of data exhibited interesting common bean landrace germplasms with the potential for use in the genetic improvement of the P. vulgaris in the northern region of Mexico.
Acknowledgments
This work was supported by the Secretaría de Investigación y Posgrado of the Instituto Politécnico Nacional (IPN) (grants 20060056, 20070061, 20080666, 20131450) and Fondo Mixto (FOMIX) del Estado de Veracruz (grant 94,070). Gill-Langarica (HRGL), R. Rosales-Serna, S. Hernandez-Delgado (SHD) and N. Mayek-Perez (NMP) are SNI fellows, and HRGL, SHD and NMP are supported by COFAA and EDI-IPN scholarships.
Bibliography
1. ACOSTA, G. J., J. S., MURUAGA, F. CÁRDENAS& M. M. KHAIRALLAH. 1996. Estrategias para la utilización de germoplasma de Phaseolus en el mejoramiento genético [Strategies for the utilization of Phaseolus germplasm in genetic improvement]. Ciência (México) 47: 149-160. [ Links ]
2. ACOSTA, J., S. GUZMÁN, G. ESQUIVEL& R. ROSALES S. 2002. El mejoramiento del frijol (Phaseolus vulgaris L.) en México: avances y perspectivas [Bean breeding (Phaseolus vulgaris L.) in Mexico: advances and perspectives], in: El fitomejoramiento ante los avances científicos y tecnológicos [Plant breeding vs. scientific and technological advances]. Memoria del Simposio. XIX Congreso Nacional de Fitogenética, Somefi, México. pp. 20-27. [ Links ]
3. ACOSTA, J. A., T. S. HERRERA, B. AGUILAR& P. GEPTS. 1999. Seed yield of segregating populations of cultivated x wild Phaseolus vulgaris. Annu. Rep. Coop. 42: 93-94. [ Links ]
4. APPA RAO, S., C. BOUNPHANOUSAY, J. M. SCHILLER, A. P. ALCANTARA& M. T. JACKSON. 2002. Naming of traditional rice varieties by farmers in the Lao PDR. Genet. Resour. Crop. Evol. 49: 83- 88. [ Links ]
5. BASSETT, M. J. 1995. The dark corona character in seed coats of common bean co-segregates with the pink flower allele vlaea. J. Am. Soc. Hort. Sci. 120: 520-522. [ Links ]
6. BEEBE, S., P. W. SKROCH, J. TOHME, M. C. DUQUE, F. PEDRAZA& J. NIENHUIS. 2000. Structure of genetic diversity among common bean landraces of middle American origin based on correspondence analysis of RAPD. Crop Sci. 40: 264-273. [ Links ]
7. BELLON, M., A. BARRIENTOS-PRIEGO, P. COLUNGA-GARCÍA MARÍN, H. PERALES, J. REYES-AGÜERO, R. ROSALES-SERNA& D. ZIZUMBO-VILLAREAL. 2009. Diversidad y conservación de recursos genéticos en plantas cultivadas [Diversity and conservation of genetic resources in crops], in: Capital natural de México. Comisión Nacional para el Conocimiento y Uso de la Biodiversidad (CONABIO), México. pp 355-382. [ Links ]
8. BITOCCHI, E., L. NANNI, E. BELLUCCI, M. ROSSI, A. GIARDINI, P. S. ZEULI, G. LOGOZZO, J. STOUGAARD, P. MCCLEAN, G. ATTENE& R. PAPA. 2012. Mesoamerican origin of the common bean (Phaseolus vulgaris L.) is revealed by sequence data. Proc. Natl. Acad. Sci. U.S.A.109: E788-E796. [ Links ]
9. BLAIR, M. W., L. M. DÍAZ, H. F. BUENDÍA & M. C. DUQUE. 2009. Genetic diversity, seed size associations and population structure of a core collection of common beans (Phaseolus vulgaris L.). Theor. Appl. Genet.119: 955-972. [ Links ]
10. BLAIR, M. W., L. M. DÍAZ, H. R. GILL-LANGARICA, R. ROSALES-SERNA, N. MAYEK-PÉREZ& J. A. ACOSTA-GALLEGOS. 2011. Genetic relatedness of Mexican common bean cultivars revealed by microsatellite markers. Crop Sci. 51: 2655-2667. [ Links ]
11. BOCZKOWSKA, M., Z. BULIŃSKA-RADOMSKA& J. NOWOSIELSKI. 2012. AFLP analysis of genetic diversity in five accessions of Polish runner bean (Phaseolus coccineus L.). Genet. Resour. Crop Evol. 59: 473-478. [ Links ]
12. BROUGHTON, W. J., G. HERNÁNDEZ, M. W. BLAIR, S. BEEBE, P. GEPTS& J. VANDERLEYDEN. 2003. Beans (Phaseolus spp.) - model food legumes. Plant Soil 252: 55-128. [ Links ]
13. BUNTJER, J. B. 1999. Cross Checker v. 2.91. Department of Plant, Breeding, Wageningen University and Research Centre, Wageningen. [ Links ]
14. CÁRDENAS, R. F. A. 1984. Clasificación preliminar de los frijoles en México [Preliminary classification of bean crops in Mexico]. Folleto Técnico No. 81. INIFAP-SARH, México. 59 p. [ Links ]
15. CÁRDENAS, R. F. A. 2000. Investigación agrícola sobre frijol en México durante el período 1943 a 1980 [Agricultural research on bean crops in Mexico during the period 1943-1980] [ Links ]
16. CÁRDENAS, R. F. A., J. S., MURUAGA-MARTÍNEZ& J. ACOSTA J. 1996. Banco de germoplasma de Phaseolus spp. del Instituto Nacional de Investigaciones Agrícolas, Forestales y Pecuarias [Germplasm bank of Phaseolus spp. at the National Institute of Forestry, Agricultural and Livestock]. CONABIO/INIFAP-Centro de Investigaciones de la Región Centro, Campo Experimental Valle de México, México, D. F. p. 421. [ Links ]
17. DEBOUCK, D. G. & R. HIDALGO. 1982. Morfología de la planta de frijol común; guía de estudio para ser usada como complemento de la unidad audiotutorial sobre el mismo tema [Morphology of common bean; study guide to be used as a complement to the audio-tutorial unit on the sametopic]. Centro Internacional de Agricultura Tropical. p. 56. [ Links ]
18. DELGADO-SALINAS, A., R. BIBLER& M. LAVIN. 2006. Phylogeny of the genus Phaseolus (Leguminosae): A recent diversification in an ancient landscape. Syst. Bot. 31: 779-791. [ Links ]
19. DELLAPORTA, S. L., J. WOODS& J. B. HICKS. 1983. A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1: 19-21. [ Links ]
20. EXCOFFIER, L. & H. E. LISCHER. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and windows. Mol. Ecol. Resour.10: 564-567. [ Links ]
21. GARCÍA, E. H., C. B. PEÑA-VALDIVIA; J. R. AGUIRRE& J. S. MURUAGA. 1997. Morphological and agronomic traits of a wild population and an improved cultivar of common bean (Phaseolus vulgaris L.). Ann. Bot. 79: 207-213. [ Links ]
22. GEPTS, P.& D. G. DEBOUCK. 1991. Origin, domestication, and evolution of the common bean (Phaseolus vulgaris L.), in: VAN SCHONHOOVEN, A. AND O. VOYSES (Eds.), Common beans: research for crop improvement. CIAT-C.A.B. International, Wallingford, Oxford, UK. pp. 7-53. [ Links ]
23. GUO, S. W.& E. A. THOMPSON. 1992. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48: 361-372. [ Links ]
24. GUPTA, S., M. SRIVASTAVA, G. P. MISHRA, P. K. NAIK, R. S. CHAUHAN& S. K. TIWARI. 2008. Analogy of ISSR and RAPD markers for comparative analysis of genetic diversity among different Jatropha curcas genotypes. Afr. J. Biotechnol. 7: 4230-4243. [ Links ]
25. HUFF, D. R., R. PEAKALL& P. E. SMOUSE. 1993. RAPD variation within and among natural-populations of outcrossing buffalo grass [Buchloe dactyloides (Nutt) engelm.].Theor. Appl. Genet. 86: 927-934. [ Links ]
26. IBPGR (INTERNATIONAL BOARD FOR PLANT GENETIC RESOURCES), 1982. Phaseolus vulgaris descriptors. Plant production and Protection Division. Rome, Italy. 32 p. [ Links ]
27. KO, H. L., D. C.COWAN, R. J.HENRY, G. C.GRAHAM, A. B. BLAKENEY& L. G. LEWIN. 1994. Random amplified polymorphic DNA analysis of Australian rice (Oryza sativa L.) varieties. Euphytica 80: 179- 189. [ Links ]
28. KUMAR, V., S. SHARMA, S. KERO, S. SHARMA, A. K. SHARMA, M. KUMAR& K. VENKATARAMANA BHAT. 2008. Assessment of genetic diversity in common bean (Phaseolus vulgaris L.) germplasm using amplified fragment length polymorphism (AFLP). Sci. Hortic. 116: 138-143. [ Links ]
29. LAURENTIN, H.& P. KARLOVSKY. 2007. AFLP fingerprinting of sesame (Sesamum indicum L.) cultivars: identification, genetic relationship and comparison of AFLP informativeness parameters. Genet. Resour. Crop Evol. 54: 1437-1446. [ Links ]
30. LÓPEZ, S. J. L., J. A. RUIZ-CORRAL, J. J. SÁNCHEZ-GONZÁLEZ& R. LÉPIZ I. 2005. Adaptación climática de 25 especies de frijol silvestre (Phaseolus spp) en la República Mexicana. Rev. Fitotec. Mex. 28: 221-230. [ Links ]
31. NEI, M.& W. H. LI. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U.S.A.76: 5269-5273. [ Links ]
32. PAPA, R.& P. GEPTS. 2003.Asymmetry of gene flow and differential geographical structure of molecular diversity in wild and domesticated common bean (Phaseolus vulgaris L.) from Mesoamerica. Theor. Appl. Genet.106: 239-250. [ Links ]
33. PECINA-QUINTERO, V., J. L. ANAYA-LÓPEZ, A. Z. COLMENERO, N. M. GARCÍA, C. A. NÚÑEZ COLÍN, J. L. SOLIS BONILLA, M. R. AGUILAR-RANGEL, H. R. GILL LANGARICA. 2011. Molecular characterisation of Jatropha curcas L. genetic resources from Chiapas, Mexico through AFLP markers. Biomass Bioenerg. 35: 1897-1905. [ Links ]
34. PERRIER, X.& C. J. JACQUEMOUD. 2006. DARwin. Software.http://darwin.cirad.fr. [ Links ]
35. KÜPPER CARDOSO PERSEGUINI, J. M., A. F. CHIORATTO, M. I. ZUCCHI, C. A. COLOMBO, S. A. CARBONELL, J. M. COSTA MONDEGO, R. GAZAFFI, A. A. FRANCO GARCIA, T. DE CAMPOS, A. P. DE SOUZA, L. B. RUBIANO. 2011. Genetic diversity in cultivated carioca common beans based on molecular marker analysis. Genet. Mol. Biol. 34: 88-102. [ Links ]
36. POWELL, W., M. MORGANTE, C. ANDRE, M. HANAFEY, J. VOGEL, S. TINGEY& A. RAFALSKI. 1996. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol. Breeding 2: 225-238. [ Links ]
37. PREVOST, A.& M. J. WILKINSON. 1999. A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor. Appl. Genet. 98: 107-112. [ Links ]
38. ROGERS, D. J.& T. T. TANIMOTO. 1960. A computer program for classifying plants. Science 132: 1115- 1118. [ Links ]
39. ROHLF, F. N. 2009. Numerical, taxonomy and multivariate analysis system (NTSYSpc) version 2.2. Exeter Software, Applied Biostatistics, Inc., New York, USA. 43 p. [ Links ]
40. ROLDAN, R. I., J. DENDAUW, E. VAN BOCKSTAELE, A. DEPICKER& M. DE LOOSE. 2000. AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol. Breeding 6: 125- 134. [ Links ]
41. ROSALES-SERNA R., R. O. MARQUEZ& J. A. A. GALLEGOS. 2001. Phenology and yield of dry bean in the Mexican highlands and its response to photoperiod. Agrociencia 35: 513-523. [ Links ]
42. ROSALES-SERNA R., S. HERNÁNDEZ-DELGADO, M. GONZÁLEZ-PAZ, J. A. ACOSTA-GALLEGOS& N. MAYEK-PÉREZ. 2005. Genetic relationships and diversity revealed by AFLP markers in Mexican common bean bred cultivars. Crop Sci. 45: 1951- 1957. [ Links ]
43. ROSALES-SERNA R., J. A. ACOSTA-GALLEGOS, R. P. DURÁN-DURÁN, H. G. ANDRADE, P. PÉREZ-HERRERA& J. S. MURUAGA-MARTINEZ. 2003. Diversidad genética del germoplasma mejorado de frijol (Phaseolus vulgaris L.) en México [Genetic diversity of improved bean germplasm (Phaseolus vulgaris L.) in Mexico]. Agric. Téc. Méx. 29: 11-24. [ Links ]
44. ROSALES-SERNA R., J. A. ACOSTA, J. S. MURUAGA, J. M. HERNÁNDEZ, G. E. ESQUIVEL& P. HERRERA. 2004. Variedades mejoradas de frijol del Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias (INIFAP) [Improved vean varieties from the National Institute of Forestry, Agriculture and Livestock (INIFAP)]. Libro Téc. 6: 148. [ Links ]
45. SNICS (Servicio Nacional de Inspección y Certificación de Semillas) [SNICS (National Seed Inspection and CertificationService]. 2001. Guía técnica para la descripción varietal de frijol. (Phaseolus vulgaris L.) [Technical Guide for the varietal description of bean].Tlalnepantla, México. 21 p. [ Links ]
46. TATIKONDA, L., S. P. WANI, S. KANNAN, N. BEERELLI, T. K. SREEDEVI& D. A. HOISINGTON, P. DEVI& R. K. VARSHNEY. 2009. AFLP-based molecular characterization of an elite germplasm collection of Jatropha curcas L., a biofuel plant. Plant Sci. 176: 505-513. [ Links ]
47. TESHOME, A., L. FAHRIG, J. K. TORRANCE, J. D. LAMBERT, T. J. ARNASON& B. R. BAUM, 1999. Maintenance of sorghum (Sorghum bicolor, Poaceae) landrace diversity by farmers' selection in Ethiopia. Econ. Bot. 53: 79-88. [ Links ]
48. VAN SCHOONHOVEN, A.& M. A. PASTOR-CORRALES, 1987. Sistema estándar para la evaluación de germoplasma de frijol [Standard system for the evaluation of bean germplasm]. Centro Internacional de Agricultura Tropical (CIAT). 56 p. [ Links ]
49. VARGAS-VÁZQUEZ, M. L. P., J. S., MURUAGA, J. A. ACOSTA , R. NAVARRETE, P. PEREZ, G. ESQUIVEL, G. IRIZAR& J. M. HERNANDEZ, 2006. Colección nucleo de Phaseolus vulgaris L. del Inifap: Catalogo de Accesiones de la Forma Domesticada [Core Collection of Phaseolus vulgaris L. at INIFAP: Catalogue of Accessions to the Domesticated Species]. Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias, Campo Experimental Valle de México, Chapingo, Edo de México. 256 p. [ Links ]
50. VARSHNEY, R. K., K. CHABANE, P. S. HENDRE, R. K. AGGARWAL& A. GRANER, 2007. Comparative assessment of EST-SSR, EST-SNP and AFLP markers for evaluation of genetic diversity and conservation of genetic resources using wild, cultivated and elite barley. Plant Sci. 173: 638-649. [ Links ]
51. VOS, P., R. HOGERS, M. BLEEKER, M. REIJANS, T. VAN DE LEE& M. HORNES, A. FRIJTERS, J. POT, J. PELEMAN& M. KUIPER. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23, 4407-4414. [ Links ]
52. VOYSEST, V. O., 2000. Mejoramiento genético del frijol (Phaseolus vulgaris L.). Legado de variedades de América Latina 1930-1999 [Bean breeding (Phaseolus vulgaris L.). Legacy of varieties of Latin America 1930-1999]. Centro Internacional de Agricultura Tropical (CIAT). Cali, Colombia. 220 p. [ Links ]
Recibido el 3 de octubre de 2013,
aceptado el 26 de junio de 2014.














