Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO  uBio
uBio
Compartilhar
Boletín de la Sociedad Argentina de Botánica
versão On-line ISSN 1851-2372
Bol. Soc. Argent. Bot. vol.49 no.4 Córdoba dez. 2014
GENÉTICA Y EVOLUCIÓN
Genome characterization of a synthetic Triticum x Thinopyrum (Poaceae) amphiploid usingin situ hybridization
Maia Fradkin1, Eduardo J. Greizerstein1, María R. Ferrari2 and Lidia Poggio3,4
1 Cátedra de Mejoramiento Genético, Facultad de Ciencias Agrarias, UNLZ, Ruta 4-Km 2, Lavallol (1836), Prov. Buenos Aires, Argentina.
2 Facultad de Ciencias Veterinarias, Universidad de Buenos Aires, Chorroarín 280 (1427), C.A.B.A., Argentina.
3 Laboratorio de Citogenética y Evolución, Instituto de Ecología, Genética y Evolución de Buenos Aires (IEGEBA) - CONICET, Intendente Güiraldes 2160 (C1428EGA), C.A.B.A, Argentina
4 Facultad de Ciencias Exactas y Naturales, Universidad de Buenos Aires, Intendente Güiraldes 2160, (C1428EGA), C.A.B.A, Argentina.
Summary
"Trigopiros" derive from crosses between different species of Triticum L. and Thinopyrum Löve. These synthetic amphiploids are designed with the aim to obtain cereals similar to wheat but which are perennial, resistant to diseases and to the salinity of the soils. Moreover, they allow the transfer of the agronomic attributes of Thinopyrum to wheat. "Trigopiro" Don Noé INTA, which is currently grown in Argentina, presents valuable agronomic traits as well as a high content of seed proteins. In the present work, the use of classical cytogenetic techniques allowed us to confirm that the chromosome number of "Trigopiro" Don Noé is 2n=56. The application of in situ hybridization (FISH-GISH) allowed us to postulate its genomic composition for the first time. This artificial hybrid has 14 chromosomes from genome J of Thinopyrum and 2 chromosomes pairs with putative translocations between Triticum and Thinopyrum. The rest of chromosomes belong to A, B and D genomes of Triticum.
Key words: "Trigopiro"; FISH; GISH.
Resumen
Caracterización del genoma de un Triticum x Thinopyrum (Poaceae) sintético amfiploide utilizando hibridación in situ. Los "Trigopiros" derivan de cruzamientos entre diferentes especies de Triticum y Thinopyrum. El objetivo es obtener cereales con características similares al trigo, perennes, resistentes a enfermedades y a la salinidad de los suelos. Además estos híbridos sintéticos son útiles para transferir al trigo atributos agronómicos de Thinopyrum. "Trigopiro" Don Noé INTA, que se cultiva actualmente en Argentina, presenta rasgos agronómicos valiosos, así como un alto contenido de proteínas seminales. En el presente trabajo se confirmó que el número de cromosomas de "Trigopiro" Don Noé es 2n = 56. Técnicas de hibridación in situ (FISH-GISH) permitieron postular su composición genómica desconocida hasta el momento. Este híbrido artificial posee 14 cromosomas del genoma J de Thinopyrum y 2 pares de cromosomas con posibles translocaciones entre Triticum y Thinopyrum. El resto de los cromosomas pertenecen a los genomas A, B y D de Triticum.
Palabras clave: "Trigopiro"; FISH; GISH.
Introduction
The name "Trigopiro" refers to cereals derived from crosses between Triticum L. and Thinopyrum Löve. These artificial amphiploids are also designated with the Latin names Agroticum, Agrotriticum, or Tritipyron (Covas et al., 1980). They were obtained with the aim to have a cereal similar to wheat but which were, resistant to diseases and to the salinity of soils. Besides, they are currently subject of cytogenetic studies because they may have structural rearrangements which cause variations in valuable agronomic traits (Han et al., 2004; Brasileiro-Vidal et al., 2005; Qi et al., 2010; Chen et al., 2012).
Thinopyrum is a relatively young genus within the tribe Triticeae that includes species with different ploidy levels. According to Dewey (1984), the species Th. elongatum (Host) D.R. Dewey, (2n=2x=14) would carry genome E whereas Th. bessarabicum (Savul. et Rayss) Á. Löve, (2n=2x=14) would carry genome J. Genomic in situ hybridization (GISH) has shown that genomes E and J are closely similar in their repetitive DNA (Kosina& Heslop-Harrison, 1996). Based on molecular cytogenetic studies, Chen et al., (1998) suggested that Th. ponticum (Podp.) Barkworth et D.R. Dewey, (2n = 10x = 70) would have a basic JJJJSJS genomic constitution and that genome JS is characterized by having specific sequences of the genome S of Pseudoroegneria strigosa (M. Bieb.) Á. Löve, in regions close to the centromere.
In Argentina, SH16 INTA and Don Noé INTA are the "trigopiros" currently used in improvement assays. In previous studies, we have found that SH16 INTA is hexaploid, with 2n=42 chromosomes, 14 of which belong to the J genome of Thinopyrum and 28 to wheat genomes. We also found that 14 of the latter belong to genome B, 4 to genome D (chromosome pairs 2D and 4D) and the remaining ones probably to genome A (Fradkin et al., 2011).
"Trigopiro" Don Noé INTA comes from an original crossing probably made at the University of California (USA) and then introduced in Argentina by the agronomist Roberto Leiboff about 45 years ago. This material, which was markedly heterogeneous, was selected at the Anguil Experimental Station of the National Institute of Agricultural Technology of Argentina (INTA) and used to obtain the variety Don Alfredo (Covas et al. 1978, 1980).
The use of genealogical selection in plants of this variety allowed obtaining a new improved variety with higher forage and grain productivity, characterized by its increased content of seminal proteins.
In this work we discuss the chromosome number and genome composition of this valuable artificial amphiploid with the aim to achieve a more efficient genetic improvement.
Materials and Methods
Plant material: Seeds of the "trigopiro" Don Noé INTA, Triticum aestivum L., var. Chinese Spring and Th. ponticum were kindly donated by Ing. Agr. G. Covas, Ing. Agr. H. Paccapelo and Prof. Ing. V. Ferreira.
Preparation of cells: Roots (1 cm long) were pretreated at 0°C in water in equilibrium with ice for 36 h, fixed in 3:1 absolute alcohol:glacial acetic acid for 24 h at room temperature, and subsequently kept at -20°C. The processing of the roots to obtain metaphase cells was carried out following Fradkin et al. (2011).
Feulgen reaction: The Feulgen staining technique was performed according to Greizerstein et al. (1997).
DNA and probes: DNA from T. aestivum and Th. ponticum and the probes of satellite DNA pSc119.2 and pAs1 were used. Genomic DNA was isolated from adult leaves of T. aestivum var. Chinese Spring and from Th. ponticum using the Wizard® Genomic DNA purification Kit (Promega), following the manufacturer's instructions with minor modifications. GISH was performed using genomic DNA from Thinopyrum labeled with biotin and unlabeled DNA from wheat as blocking DNA (1:60). FISH was performed with the probes pSc119.2 and pAs1. The probes were labeled using the BioNick Labeling System (Invitrogen) following the manufacturer's instructions. All the probes used were revealed with streptavidin Cy3. In situ hybridization was performed following Ferrari et al. (2005), with minor modifications.
Results
Observations done on mitotic cells stained with the Feulgen Reaction (n=25), revealed a chromosome number 2n = 56 (Fig. 1a).
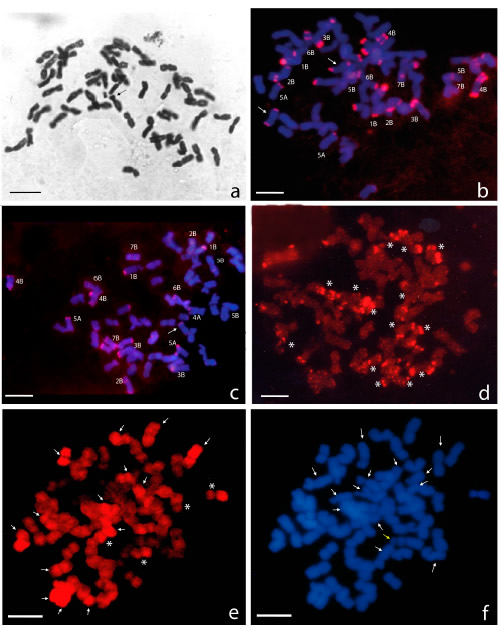
Fig. 1. Mitotic metaphase of "trigopiro" Don Noé INTA. a: Feulgen reaction. A total of 56 chromosomes are observed. The arrow indicates the 6B chromosome. b, c: FISH with pSc119.2 from Secale cereale (red). The photomicrograph shows 14 chromosomes of the B genome of wheat and the pairs 4A and 5A of wheat. The arrows correspond to chromosomes of J genome. d: FISH with pAs1 of Aegilops squarrosa (red). The asterisks indicate the presence of 14 chromosomes of the wheat D genome. e: GISH: the arrows show 14 chromosomes with intense hybridization signal with total genomic DNA from Thinopyrum ponticum (red) and the asterisk indicate putative translocation Triticum- Thinopyrum. f: Same cell as (e), counterstained with DAPI. Arrows show chromosomes with null or almost null hybridization signal with total genomic DNA from Thinopyrum ponticum. Yellow arrow points 6B chromosome. All the probes were labeled with biotin and revealed with Cy3. Bars 10 µm.
The use of pSc119.2 allowed us to observe until 28 chromosomes with hybridization signals (n=25). We recognized 18 chromosomes of wheat: 14 of the B genome and the pairs 4A and 5A. The chromosomes 2B, 4B, 5B y 6B showed interstitial signals and the remaining labeled chromosomes presented only terminal signals (Fig. 1 b and c).
FISH experiments, using pAs1, revealed chromosomes of the D genome of wheat and other ones, that based on their small size and the distribution of their hybridization signals, would belong to Thinopyrum genome (n=10) (Fig. 1 d).
In the current work, genomic DNA of Th. ponticum as a probe and DNA of wheat as a block were applied to mitotic cells (n=10), strong hybridization signals were observed in 14 chromosomes. Two chromosome pairs had an arm intensely colored and the other lightly colored. The remaining chromosomes showed light or null coloration (Fig. 1 e). DAPI counterstaining allowed us to observe clearly all the chromosomes in each cell (Fig.1 f).
Discussion
"Trigopiro" Don Noé, a cultivar with a high content of seminal proteins and which is used to obtain improved lines of wheat and synthetic hybrids within the tribe Triticeae (Covas et al., 1980; Ferreira et al., 2007; Ruiz et al., 2007; Castro et al., 2011). Thus, it is important to know its genome composition to establish strategies for its use in breeding programs.
The chromosome number determined in the present work for "trigopiro" Don Noé INTA was 2n=56, which confirms the observations made by other authors (Ruiz et al., 2000; Tosso et al., 2000).
FISH experiments using different probes enable to know the genome composition and to recognize chromosomes in diverse species and in natural and artificial hybrids. The probe designated pSc119.2, which contains a 120-bp highly repeated sequence (Bedbrook et al., 1980), was isolated from Secale cereale and thus allows identifying the rye chromosomes. Besides, it is characterized by the ability to hybridize with various species of the tribe Triticeae (Heslop-Harrison, 2000). This probe identifies the complete B genome, the pairs 4A and 5A and some chromosomes of D genome of Triticum (Mukai et al., 1993). Also gives signals in Thinopyrum and discriminates between E and J genomes. Hybridization sites are distributed throughout genome E chromosomes and at the terminal regions of the genome J chromosomes (Lapitan et al., 1987; Kosina& Heslop-Harrison 1996; Brasileiro-Vidal et al.; 2003, Sepsi et al., 2008).
In the present work, using the probe pSc119.2 in mitotic cells of "trigopiro" Don Noé, we recognized the seven chromosomes pairs of the wheat B genome and the pairs 4A and 5A. The remaining labeled chromosomes showed only terminal signals, hence they would belong to the genomes J of Thinopyrum and D of wheat.
For a more complete analysis of the genome composition of "trigopiro" Don Noé INTA, FISH experiments using pAs1 as a probe were made. This probe was isolated from the D genome of Aegilops squarrosa auct. non L. = Ae. tauschii Coss, (Rayburn& Gill, 1986). In the variety Chinese Spring, in situ hybridization with this probe allows discriminating the seven chromosomes of the D genome (Mukai et al., 1993). FISH experiments using this probe reveals chromosomes of the D genome of "trigopiro" Don Noé INTA and other chromosomes that based on their small size and their hibridization signals distribution would belong to Thinopyrum genome.
In situ hybridization with genomic DNA (GISH) has been previously used to determine the genome composition of many interspecific hybrids, as well as to detect the presence of introgressions and translocations (Ferrari et al., 2005; Wang et al., 2005; Zheng et al., 2006; Chen et al., 1998; Qi et al., 2010).
In the present work, GISH using Th. ponticum as a probe and T. aestivum as blocking allowed us to recognize 14 Thinopyrum chromosomes with a bright hybridization and two pairs of Triticum- Thinopyrum translocated chromosomes. The rest of wheat chromosomes had different intensity of hybridization indicative of different levels of cross hybridization. According to Liu et al. (2007) genetic relationships among J Thinopyrum genome and the wheat A, B, and D genomes present differences, the J genome is closer to the D genome than to either the A or B genomes.
The presence of two pairs of chromosomes with hybridization signal in only one arm suggest that translocations Triticum-Thinopyrum would occurred during the breeding process of "trigopiro" Don Noé. Several authors have described the presence of translocations between wheat and Thinopyrum chromosomes (Zhang et al., 1996; Brasileiro-Vidal et al., 2005; Oliver et al., 2006).
The cytogenetical results obtained in "trigopiro" Don Noé INTA showed important differences from those obtained in "trigopiro" SH16 INTA (Fradkin et al., 2011). Both hybrids currently used in breeding programs, present different levels of ploidy and different chromosome and genome composition.
Summarizing, "trigopiro" Don Noé INTA is a synthetic amphiploid with 2n=56 chromosomes that has 14 chromosomes from of genome J of Thinopyrum, 2 chromosomes pairs have putative translocations between Triticum and Thinopyrum and the rest of chromosomes belong to A, B and D genomes of Triticum.
Acknowledgements
The authors thank Prof. Ing. Agr. H Paccapelo and Prof. Ing V. Ferreira for kindly providing "trigopiro" Don Noé INTA seeds. This research was carried out in Argentina and supported by grants from the "Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET)" (PIP 0342), Agencia Nacional de Promoción Científica y Tecnológica (PICT bicentenario 2010-1665) and Universidad de Buenos Aires (UBACyT: 20020100100859).
Bibliography
1. BEDBROOK, J.R., J. JONES, M. O´DELL, R.D. THOMPSON,& R.B. FLAVELL. 1980. A molecular description of telomeric heterochromatin in Secale species. Cell 19: 545-560. [ Links ]
2. BRASILEIRO-VIDAL A.C., A. CUADRADO, S.P. BRAMMER, A.M. BENKO-ISEPPON& M. GUERRA. 2005. Molecular cytogenetic characterization of parental genomes in the partial amphiploid Triticum aestivum x Thinopyrum ponticum. Genet. Mol. Biol. 28: 308-313. [ Links ]
3. BRASILEIRO-VIDAL A.C., A. CUADRADO, S.P. BRAMMER, A.C. ZANNATTA, A.M. PRESTES, M.I.B. MORAES-FERNANDES& M. GUERRA. 2003. Chromosome characterization in Thinopyrum ponticum (Triticeae, Poaceae) using in situ hybridization with different DNA sequences. Genet. Mol. Biol. 26: 505-510. [ Links ]
4. CASTRO N., H. RUFACH, F. CAPELLINO, R. DOMÍNGUEZ& H. PACCAPELO 2011. Evaluación del rendimiento de forraje y grano de triticales y tricepiros. Revista Investig. Agropecu. 37: 1-9. [ Links ]
5. CHEN G., Q. ZHENG, Y. BAO, S. LIU, H. WANG& X. LI. 2012. Molecular cytogenetic identification of a novel dwarf wheat line with introgressed Thinopyrum ponticum chromatin. J. Biosci. 37: 149-55. [ Links ]
6. CHEN Q., R.L. CONNER, A. LAROCHE& J.B. THOMAS. 1998. Genome analysis of Thinopyrum intermedium and Th. ponticum using genomic in situ hybridization. Genome 41: 580-586. [ Links ]
7. COVAS G., M.A. FRECENTESEM& H. VOLONTERI. 1980. Contenido de proteína del grano de un cereal sintético trigopiro INTA. Inform. Tecnol. Agrop. Estac. Exp. Reg. Agrop. Anguil. 75: 39-42. [ Links ]
8. COVAS G., D. MONTAÑO& J. SAN MIGUEL. 1978. Trigopiro, un cereal sintético de cualidades interesantes. Inform. Tecnol. Agrop. Estac. Exp. Reg. Agrop. Anguil 73:2. [ Links ]
9. DEWEY D. R. 1984. The genomic system of classification as a guide to intergeneric hybridization with the perennial Triticeae. In: GUSTAFSON J.P. (ed) Gene Manipulation in Plant Improvement, pp. 209- 279. Plenum Publishing Corp., N.Y. [ Links ]
10. FERRARI M.R., E.J. GREIZERSTEIN, H. PACCAPELO, C. NARANJO, A. CUADRADO, N. JOUVE& L. POGGIO. 2005. The genomic composition of tricepiro, a synthetic forage crop. Genome 48: 154-159. [ Links ]
11. FERREIRA V., M. SCALDAFERRO, E. GRASSI& B. SZPINIAK. 2007. Nivel de ploidía, estabilidad citológica y fertilidad en cruzas de triticale x trigopiro (tricepiros). J. Basic Appl. Genet. 18: 15- 26. [ Links ]
12. FRADKIN M., M.R. FERRARI, V. FERREIRA, E. GRASSI, E.J. GREIZERSTEIN& L. POGGIO. 2011. Chromosome and genome composition of a Triticum x Thinopyrum hybrid by classical and molecular cytogenetic techniques. Genet. Resour. Crop Evol. 59: 231-237. [ Links ]
13. FRADKIN M., E.J. GREIZERSTEIN, H. PACCAPELO, V. FERREIRA, E. GRASSI, L. POGGIO& M.R. FERRARI. 2009. Cytological analysis of hybrids among triticales and trigopiros. Genet. Mol. Biol. 32:797-801. [ Links ]
14. GREIZERSTEIN E.J., C.A. NARANJO& L. POGGIO. 1997. Karyological studies in five wild species of Amaranths. Cytologia 62: 115-120. [ Links ]
15. HAN F., B. LIU, G. FEDAK& Z. LIU. 2004. Genomic constitution and variation in five partial amphiploids of wheat-Thinopyrum intermedium as revealed by GISH, multicolor GISH and seed storage protein analysis. Theor. Appl. Genet. 109: 1070-1076. [ Links ]
15. HESLOP-HARRISON J.S. 2000. Comparative genome organization in plants: from sequence and markers to chromatin and chromosomes. Plant Cell 12: 617-635. [ Links ]
16. KOSINA R.& J.S. HESLOP-HARRISON. 1996. Molecular cytogenetics of an amphiploid trigeneric hybrid between Triticum durum, Thinopyrum distichum and Lophopyrum elongatum. Ann. Bot. 78: 583-589. [ Links ]
17. LAPITAN N.L.V., B.S. GILL& R.G. SEARS. 1987. Genomic and phylogenetic relationships among rye and perennial species in the Triticeae. Crop Sci. 27: 682-687. [ Links ]
18. LÖVE A. 1984. Conspectus of the Triticeae. Feddes Repert. 95: 425-521. [ Links ]
19. MUKAI Y, Y. NAKAHARA& M. YAMAMOTO. 1993. Simultaneous discrimination of the three genomes in hexaploid wheat by multicolor fluorescence in situ hybridization using total genomic and highly repeated DNA probes. Genome 36: 489-495. [ Links ]
20. OLIVER R.E., S.S. XU, R.W. STACK, T.L. FRIESEN, Y. JIN& X. CAI. 2006. Molecular cytogenetic characterization of four partial wheat-Thinopyrum ponticum amphiploids and their reaction to Fusarium head blight, tan spot, and Stragonospora nodorum blotch. Theor. Appl. Genet. 112: 1473-1479. [ Links ]
21. QI Z, P.DU, B. QIAN, L. ZHUANG, H. CHEN, T. CHEN, J. SHEN, J. GUO, Y. FENG& Z. PEI. 2010. Characterization of a wheat-Thinopyrum bessarabicum (T2JS-2BS.2BL) translocation line. Theor. Appl. Genet. 121: 589-597. [ Links ]
22. RAYBURN A.L.& B.S. GILL. 1986. Molecular identification of D-genome chromosomes of wheat. Heredity 77: 253-255. [ Links ]
23. RUIZ M., H. PACCAPELO& G. COVAS. 2000. Tricepiro: cultivo y usos. Forrajes& Granos Journal 57: 50-52. [ Links ]
24. RUIZ M.A., A.D. GOLBERG& O. MARTÍNEZ. 2007. Limitación hídrica y producción de forraje y semilla de variedades de tricepiro, triticale y trigopiro. RAPA 27, Supl. 1: 188-189. [ Links ]
25. SEPSI A., I. MOLNÁR, D. SZALAY& M. MOLNÁR-LÁNG. 2008. Characterization of a leaf rust-resistant wheat-Thinopyrum ponticum partial amphiploid BE-1, using sequential multicolor GISH and FISH. Theor. Appl. Genet. 116: 825-834. [ Links ]
26. TOSSO H., H.A. PACCAPELO& G.F. COVAS. 2000. Caracterización de líneas avanzadas de Tricepiro. I. Descripción morfológica y citológica. Revista Investig. Agropecu. 29: 39-51. [ Links ]
27. WANG J., F. XIANG& G. XIA 2005. Agropyron elongatum chromatin localization on the wheat chromosomes in an introgression line. Planta 221: 277-286. [ Links ]
28. ZHANG X., Y. DONG & R. WANG. 1996. Characterization of genomes and chromosomes in partial amphiploids of the hybrid Triticum aestivum x Thinopyrum ponticum by in situ hybridization, isozyme analysis, and RAPD. Genome 39:1062- 1071. [ Links ]
29. ZHENG Q., B. LI, X. ZHANG, S. MU, H. ZHOU& H.Z. LI. 2006. Molecular cytogenetic characterization of wheat-Thinopyrum ponticum translocations bearing blue-grained gene(s) induced by r-ray. Euphytica 152: 51-60. [ Links ]
Recibido el 21 de marzo de 2014,
aceptado el 20 de mayo de 2014.














