Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
Boletín de la Sociedad Argentina de Botánica
versión On-line ISSN 1851-2372
Bol. Soc. Argent. Bot. vol.51 no.2 Córdoba jun. 2016
GENÉTICA Y EVOLUCIÓN
Chromosome differentiation in three species of Leptostemonum (Solanum, Solanaceae) endemic to oceanic islands
Franco E. Chiarini1,* and Martha J. Gauthier2
1 Instituto Multidisciplinario de Biología Vegetal (IMBIV), CONICET and Universidad Nacional de Córdoba.CC. 495, 5000, Córdoba - Argentina.
2 University of Hawaii at Manoa. Dept. of Tropical Plant and Soil Sciences, 3190 Maile Way, St. John 102, Honolulu HI 96822
*Corresponding author: E-mail: franco.e.chiarini@gmail.com
Summary
The chromosomes of Solanum nelsonii, S. sandwicense (endemic to the Hawaii islands) and S. vespertilio (from the Canary islands) were studied by means of classical staining, CMA/DAPI banding and FISH with probes for the 18-5.8-26S and 5S rDNA genes. The aim of this study was to test for chromosomal changes (chromosome number, karyotype, heterochromatin pattern, rDNA loci) during the evolution of these taxa with respect to their continental relatives. An apparent chromosome stasis was confirmed in all three species, in regards to chromosome number and karyotype morphology. However, there was also evidence of cryptic, cumulative sequence changes. Speciation in these species is not likely associated with large, obvious chromosome rearrangements or polyploidy, but more likely due to genetic divergence.
Key words: Chromosome stasis; Heterochromatin; Ribosomal DNA; Solanum subgen. Leptostemonum.
Resumen
Diferenciación cromosómica en tres especies de Leptostemonum (Solanum, Solanaceae) endémicos de islas oceánicas.
Los cromosomas de Solanum nelsonii, S. sandwicense (endémicos de las islas Hawaii) y de S. vespertilio (de las islas Canarias) fueron estudiados por medio de tinción clásica, bandeo CMA/DAPI y FISH con sondas para los genes de ADNr de 18-5.8-26S y 5S. El objetivo fue comprobar cambios cromosómicos (número cromosómico, cariotipo, patrones de heterocromatina y loci de ADNr) durante la evolución de estos taxones, en comparación con sus parientes continentales. Una aparente estasis cromosómica fue confirmada para las tres especies, en lo que respecta a número cromosómico y cariotipos. Sin embargo, también se encontró evidencia de cambios de secuencias crípticos, acumulativos. La especiación en estas especies no estaría asociada a grandes rearreglos cromosómicos o poliploidía, sino más bien a divergencia genética.
Palabras clave: Estasis cromosómica; Heterocromatina; ADN ribosómico; Solanum subgen. Leptostemonum.
Introduction
Oceanic islands provide a special case for studying the processes and results of evolution (e.g. Vitousek et al., 1995; Stuessy & Ono, 1998), since their temporal boundaries (datable volcanic eruptions) and geographical borders (unfavourable salt water) are well defined. When the arrival of species can be dated precisely, differences between these taxa and their continental relatives can be evaluated more easily than with other organisms. For this reason, taxa from the Hawaiian and Canary archipelagos have been the focus of much evolutionary based research (Wagner & Funk, 1995; Bramwell & Caujapé-Castells, 2011). Island floras are unique as compared to their mainland counterparts and are characterized by an impoverishment and disharmony: some taxa are overrepresented, some are underrepresented, and evolutionary rates are higher relative to mainland sources (Carlquist, 1974; Bramwell & Caujapé- Castells, 2011). In this regard, it is worth mentioning Solanaceae, a family with 2300-2700 species that is poorly represented on islands (Hunziker, 2001; Särkinen et al., 2013). This family includes Solanum L., one of the five largest genera of Angiosperms, with at least 1,250 species worldwide (Hunziker, 2001; Bohs, 2005; Särkinen et al., 2013). One third of these Solanum species belong to the subgenus Leptostemonum (the clade of the "spiny solanums"), which includes both edible plants (S. melongena L., "eggplant"; S. quitoense Lam.,"lulo") and weed species (S. elaeagnifolium Cav., "silverleaf nightshade")(Levin et al., 2006; Vorontsova et al., 2013). Despite the abundance and diversity of the family, there are just 16 species of Solanaceae in the Hawaiian islands, three of which are endemic and fall under Leptostemonum (S. incompletum Dunal, S. nelsonii Dunal and S. sandwicense Hook. et Arn.); while in the Canary Islands, there are only four endemic Solanaceae, two of which belong to Leptostemonum (S. vespertilio Aiton and S. lidii Sunding). The existence of these endemic Leptostemonum spp. within two distantly separated archipelagos offers an opportunity to make comparisons and test hypothesis about their evolutionary pathways. In this study, we examined two species from the Hawaiian Islands (S. sandwicense and S.nelsonii), and S. vespertilio from the Canary Islands.
The very rare S. sandwicense ("pōpoloʻaiakeakua" or "Hawaii horsenettle") is an erect shrub, known only from a few sites in mesic forests from about 2500 to 4000 feet, on Kaua'I and O'ahu (Wagner et al., 2005). Whalen (1984) placed this species within the "Sandwicense group" (together with S. incompletum and S. kauaiense Hillebr.), while molecular phylogenies demonstrate a relationship between this species and S. incompletum, S. furfuraceum R.Br., S. stelligerum Sm., S. cinereum R.Br., S. ferocissimum Lindl., S. chenopodinum F.Muell. and S. pancheri Guillaumin (Australian species)(Bohs, 2005; Levin et al., 2006).
Solanum nelsonii (pōpolo) is a sprawling shrub that inhabits coastal dunes on several of the northwestern islands (Ni'ihau, Nihoa, Midway Islands). This species used to be quite common in these areas, but is now increasingly difficult to find and restricted to a few sites that have evaded the negative impacts of human activity. The taxonomic position of S. nelsonii within Leptostemonum remains uncertain as it has not been included in any molecular phylogenetic studies.
Solanum vespertilio (or "rejalgadera") is a shrub, which exists within a single population of three individuals on Gran Canaria and about seven scattered relictual populations along forest margins on Tenerife composed of about 230 individuals (Anderson et al., 2006; Bramwell, 2006). According to Bohs (2005) and Weese & Bohs (2007) it is close to African species (S. melongena, S. macrocarpon L., S. aethiopicum L.), while in the phylogeny of Levin et al., (2006) it is part of the "Canary clade", allied to African species (E.g. S. aethiopicum, S. anguivi Lam., S. capense L.).
Surveys of cytological data from oceanic islands reveal little chromosomal change during the evolution of endemic plants, even though speciation is often accompanied by remarkable morphological differentiation (e.g. in Dendroseris, Stuessy & Ono, 1998; in Cyanea, Givnish et al., 2009). Stasis of chromosome number during speciation is another notable feature attributed to the evolution of oceanic island angiosperms (Sanders et al., 1983; Stuessy & Crawford, 1998; Carr, 1998). Explanations for this phenomenon include low levels of hybridization due to reproductive isolation, short periods of geological time and selection against cytotypes which might alter the adaptive traits that permit successful colonization and radiation. Considering this, the aim of this work was to look for chromosomal changes (chromosome number, karyotype, heterochromatin pattern, rDNA loci) during the evolution of S. sandwicense, S.nelsonii and S. vespertilio respective to their continental relatives.
Material and Methods
Voucher specimens.
The provenance of the plant material studied in Solanum vespertilio is: Spain, Canary Islands, Tenerife, Las Bodegas, Anderson and Santos Guerra 4601, 01/January/2004 (ORT, CONN).
In the case of S. nelsonii and S. sandwicense, rootlets were extracted from plants purchased at ‘Hui Ku Maoli Ola' plant nursery on Oahu, HI.
Chromosome preparations
Mitotic chromosomes were examined in root tips obtained from germinating seeds (S. vespertilio) or from rootlets rootlets that were extracted from adult individuals without causing harm (S. nelsonii and S. sandwicense). Roots were pretreated in saturated p-dichlorobenzene in water for 2 h at room temperature, fixed in 3:1 ethanol:acetic acid, washed in distilled water, digested 45 min at 37° C with Pectinex SP ULTRA® (Novozymes), and squashed in a drop of 45% acetic acid. After coverslip removal in liquid nitrogen, the slides were stored at –20 ° C. For mitotic counts and karyotypes, slides were stained with Giemsa (Guerra, 1983). At least ten metaphases of each species were photographed with phase contrast in a Zeiss Axiophot microscope. Photographs were used to take the following measurements for each chromosome pair: s (short arm), l (long arm), and c (total chromosome length). The arm ratio (r=l/s) was then calculated and used to classify the chromosomes as recognized by Levan et al., (1964). In addition, total haploid chromosome length of the karyotype (tl) based on the mean chromosome lengths was calculated. Karyotype asymmetry was estimated using Romero Zarco's (1986) indices (A1 = intrachromosomal asymmetry index, and A2 = interchromosomal asymmetry index). Idiograms were based on the mean values for each species.
Karyotypes and CMA/DAPI banding
Slides were stained with a drop of 0.5 mg/ml chromomycin A3 (CMA) in McIlvaine buffer, pH 7.0 and distilled water (1: 1) containing 2.5 mM MgCl2 for 90 min and subsequently stained with 2 mg/ml 4'-6-diamidino-2-phenylindole (DAPI) for 30 min, and finally mounted in McIlvaine's buffer-glycerol v/v 1: 1 (Schweizer, 1976; Schweizer & Ambros, 1994). The amount of heterochromatin was expressed as a percentage of the total length of the haploid karyotype.
Fluorescent in situ hybridization (FISH)
The location and number of rDNA sites were determined by FISH using the following probes: the pTa71 containing the 18-5.8-26S gene of wheat (Gerlach & Bedbrook, 1979) labeled with biotin-14-dATP (BioNick, Invitrogen Carlsbad); a 5S rDNA fragment obtained by PCR (Kitamura et al., 2001) with DNA of S. stuckertii, labeled with digoxigenin-11-dUTP (DigNick, Roche). The FISH protocol was according to Schwarzacher & Heslop-Harrison (2000), with minor modifications. The preparations were incubated in 100mg/ml RNAse, post-fixed in 4% (w/v) paraformaldehyde, dehydrated in a 70–100% graded ethanol series, and air-dried. On each slide 15ml of hybridization mixture was added (4–6 ng/ml of probe, 50% formamide, 10% dextran sulfate, 2x SSC and 0.3% SDS), previously denatured at 70 ° C for 10 min. Chromosome denaturation/hybridization was done at 90° C for 10 min, 48° C for 10 min, and 38° C for 5 min using a thermal cycler (Mastercycler, Eppendorf, Hamburg, Germany), and slides were placed in a humid chamber at 37° C overnight. The 18-5.8-26S probe was detected with avidin- FITC conjugate (Sigma-Aldrich), the 5S probe was detected with antidigoxigenin-rhodamine (Roche), and then counterstained and mounted with 25ml antifade Vectashield ® (Vector Lab.), containing 1.5 mg/ml of DAPI. Photomicrography was done with a Zeiss Axiophot microscope equipped with epifluorescence and a digital image capture system. For the mergence of images the free software ImageJ was employed (http://rsbweb.nih.gov/ij/).
For each species, some of the FISH slides were discolored and then stained with CMA/DAPI to study the co-localization of rDNA signals and heterochromatic bands.
Results
Chromosome numbers and karyotypes
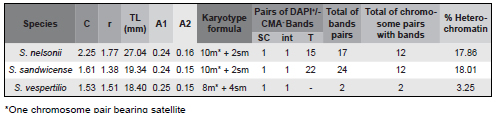
Chromosomes were found to be relatively small (Fig. 1, 2; Table 1). The chromosome traits analyzed are summarized in Table 1 and displayed schematically in Fig. 2. Accessions for all three species presented 2n = 24. All species showed one satellite placed in the short arm of the largest m chromosome of the complement. Karyotypes were quite symmetric, with the majority of formulae composed of m chromosomes.

Fig. 1. Mitotic metaphases of Solanum stained with CMA (left column) and detected by FISH with the 18- 5.8-26S and 5S probes (right column). a-b, Solanum nelsonii; c-d, S. sandwicense; e-f, S. vespertilio. Arrow heads point to chromosomes bearing the CMA+NOR co-localized with the 18-5.8-26S signal; Arrows point to chromosomes with the CMA+ band corresponding to the 5S site; asterisks point to the chromosome with the 5S site. All at the same scale. In e, the lower right box shows the heteromorphic chromosome found in one individual of S. vespertilio.
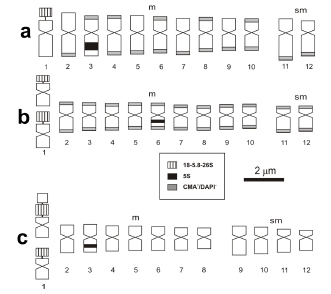
Fig. 2. Idiograms of Solanum species based on mean chromosome values, all at the same scale. a, Solanum nelsonii; b, S. sandwicense; c, S. vespertilio. Chromosomes are ordered from longest to shortest within each category, from m to sm. Grey blocks indicate CMA+/DAPI– bands, black blocks are 5S signals and hatched blocks are 18-5.8-26S signals plus CMA+/DAPI– band in the same position. In S. sandwicense and S. vespertilio both chromosomes of the heteromorphic pairs are showed.
Table 1. Cytogenetic features in the three Solanum species analyzed: Karyotype variables, number of FISH signals, heterochromatic bands and percentage of heterochromatin. C= average chromosome length, r = arm ratio, TL = total haploid chromosome length of the karyotype, A1 = intrachromosomal asymmetry index, and A2 = interchromosomal asymmetry index; SC = secondary constriction, int = intercalary band; T = terminal band.
CMA/DAPI banding
The chromosome banding showed three different heterochromatin types: (1) the three species presented a strong pair of CMA signals, corresponding to GC-rich heterochromatin regions, associated with the secondary constrictions (NOR) in terminal position (Fig. 1, 2); (2) in Solanum nelsonii and S. sandwicense, (Fig. 1a-b, Fig. 1c-d, respectively), additional CMA+/ DAPI- heterochromatin blocks not associated with NOR were located in terminal regions, with an almost equilocal distribution between non-homologous chromosomes of the karyotype; (3) In the three species, one CMA+/DAPI-interstitial band was observed in one of the pairs (Fig. 1, 2), corresponding to the 5S signal. The total amount of the CMA+ heterochromatin ranged from 3.25 to 18.01 % of the total karyotype length (Table 1).
Fluorescent in situ hybridization (FISH)
Chromosomes bearing one secondary constriction are obvious in the three species and are strongly marked with FISH using 18-5.8-26S rDNA probes. All species showed two terminal sites (Fig. 1, 2; Table 1), which coincide with a CMA+/DAPI- block. The morphology of NOR-bearing chromosomes and the localization of the 18-5.8- 26S rDNA loci was the same for the three species: the signal was located in an m chromosome, the largest of the complement. Solanum sandwicense showed a heteromorphic satellite pair, with one of the 18-5.8-26S blocks smaller than the other (Fig. 1c, d). In one individual of S. vespertilio heteromorphism was observed in the NOR bearing pair. In this case, one of the homologs presented an extra constriction after the NOR, consisting of a euchromatic chromosomal segment in terminal position (Fig. 1e).
The hybridization signals obtained with the 5S rDNA probe were two for each species (Fig. 1, 2). The position of these signals was subterminal and/ or interstitial and placed in the long arm, in an m chromosome (Fig. 1). In all cases, the 5S sites are non-syntenic with respect to the 18-5.8-26S.
Discussion
Chromosome numbers and karyotype features
This is the first chromosome number report for S. sandwicense, while the numbers found for S. nelsonii and S. vespertilio confirm previous reports (Carr, 1985; Chiarini et al., 2006). The subfamiliy Solanoideae, in which these study species are included, is also recognized as the "X=12 clade", as these numbers strongly predominate among all genera within this subfamily (Olmstead et al., 2008). Solanum is no exception and most of these species are diploids with 2n=24, with a few exceptions (Chiarini & Bernardello, 2006). Thus, no alterations on chromosome number or ploidy level took place during the speciation process of these Solanum species in the Hawaiian and Canary islands. Despite the great distance separating the Canary from the Hawaiian islands (and the distance of both archipelagos to the continent), these three study species share a single chromosomal pattern with the only continental Solanum spp. analyzed (Acosta et al., 2005; Chiarini & Bernardello, 2006; Melo et al., 2011; Moyetta et al., 2013). The cytogenetic features of these study species are within the overall pattern recognized within the Solanum genus; chromosomes are very small (1-2 mm), karyotypes are mostly composed by m chromosomes, asymmetry indexes are relatively low, and one chromosome pair bears satellites (usually the largest of the complement). An apparent chromosome stasis is confirmed in S. nelsonii, S. sandwicense and S. vespertilio, at least in regards to chromosome number and karyotype morphology.
CMA/DAPI banding
Unlike S. vespertilio, both S. nelsonii and S. sandwicense exhibited a marked number of CMA+/ DAPI- bands. Heterochromatin amount may vary considerably, even among closely related species, such as Solanum sect. Acanthophora spp. (Rego et al., 2009; Melo et al., 2011; Chiarini et al., 2014) and Capsicum spp. (Scaldaferro et al., 2013). It is hypothesized that this is an extra component of the genome, because in most Solanaceae species analyzed, constitutive heterochromatin amount is positively correlated with karyotype length and genome size, which is consistent with our findings (Moscone et al., 1996, 2007; Chiarini et al., 2014).
Highly GC-rich heterochromatin is common in Solanum spp. (Rego et al., 2009; Melo et al., 2011; Chiarini et al., 2014), occurring at macrosatellites, and at a variable number of other distal bands and intercalary bands. NOR-associated heterochromatin of the Solanum species studied here, containing the 18-5.8-26S genes, are GC rich, as is the general rule in plant chromosomes (Guerra, 2000; Scaldaferro et al., 2013; Garcia et al., 2013).
In the species analyzed, the different types of heterochromatin occupy similar positions of non-homologous chromosomes of the karyotype. Cytological evidence in several organisms showed that this pattern is probably caused by unequal crossing-over or amplification and transposition events between them mediated by Rabl orientation during mitotic interphase or bouquet arrangement during meiotic prophase (Dover & Flavell, 1984; Schweizer & Loidl 1987; Guerra, 2000). In Solanum the different classes of heterochromatin are likely made up of similar tandem repeats, displaying almost equilocal distribution between non-homologous chromosomes of the karyotype, which suggests concerted evolution for the heterochromatin dispersion in the genus.
The pattern of terminal heterochromatic bands in most or all chromosomes of the complement seems to be a homoplasic character that have appeared several times independently within Leptostemonum. Such pattern was observed in four species of the Torva clade (Rego et al. 2009), in one of Acanthophora (Chiarini et al. 2014), in S. elaeagnifolium and in S. sisymbriifolium (Acosta et al. 2013). Species with heterochromatic bands restricted only to NORs have been reported in Acanthophora, Melongena and Torva clades (Rego et al. 2009; Melo et al. 2011; Chiarini et al. 2014). Therefore, this character can only be used for analyzing closely related species within a phylogeny. Thus, the pattern of S. nelsonii can not be compared to other species because its phylogenetic position within Leptostemonum is uncertain, while for S. sandwicense, although its position is known, it can not be properly discussed because no chromosomal data are available for close relatives. However, the evolution of heterochromatin patterns can be discussed in the case of S. vespertilio, which would be placed in the same clade with two continental species: S. gilo and S. melongena (Levin et al. 2006). All the three species have heterochromatic bands restricted to NORs, which reaffirm their relationship, and allows to speculate that colonization of the Canary islands have occurred without increasing of heterochromatic bands. Nevertheless, it is necessary to know the pattern in the remaining species of the clade to stand it with certainty.
Fluorescent in situ hybridization (FISH)
According to Roa & Guerra (2012) and Garcia et al., (2013), the most common rDNA pattern found in plants consists of one or two pairs of 18-5.8- 26S sites per diploid karyotype, usually located in terminal regions of short chromosome arms. Despite the wide dispersion capacity of these sequences, there could be a tendency for the number of rDNA sites to be restricted. The number of two 18-5.8- 26S sites per complement always associates with NORs satellites and seems to be the rule within the "spiny Solanums" (Rego et al., 2009; Melo et al., 2011; Chiarini et al., 2014). The results found here are consistent with this trend. However, the number of 18-5.8-26S sites seems to be variable outside of the Leptostemonum clade. For example, more than one pair occurs in S. corymbiflorum (Sendtn.) Bohs from the Cyphomandropsis clade, and in other related genera of Solanaceae from the so-called "x = 12 clade", such as Vassobia breviflora (Sendtn.) Hunz. (Rego et al., 2009). There is evidence that number and position of rDNA sites is not a fixed character, but a target of dynamic, plastic changes, as demonstrated in species of several plant families (Garcia et al., 2009; Malinska et al., 2010) and even within Solanum subgen. Leptostemonum (Chiarini et al., 2014).
It is said that several factors may be working together to limit chromosomal changes during speciation in oceanic islands (Carr, 1998; Stuessy & Ono, 1998; Weiss et al., 2002). Low levels of genetic variation within island species may lower the probability of chromosomal alterations. Further, new chromosomal rearrangements would have a low probability of becoming fixed in populations or having a negative impact in the survival of newly originated species. However, we found some obvious structural variability in S. sandwicense and S. vespertilio (i.e. heteromorphism in the chromosome pair bearing the 18-5.8-26S). In fact, Prohens et al. (2007) found relatively high genetic diversity in S. vespertilio, probably due to the combination of the many unusual reproductive features (andromonoecy, zygomorphy, heteranthery and weak enantiostyly). The presence of heteromorphism in the satellite chromosome of S. vespertilio may be due to chromosomal aberration in this particular individual (an inversion or translocation). To test for Mendelian inheritance, a broad scope chromosomal analysis would be required and would need to include homozygous individuals, or, determine if fixed heterozygotes were the result of some special reproductive system.
Despite lack of major karyological changes, it is obvious that genetic alterations accumulate during speciation of island endemics (particularly S. sandwicense and S. nelsonii) because they are morphologically distinct from each other (Weiss et al., 2002). Transposition and loss of DNA sequences could have occurred during genome evolution and island colonization or establishment without major structural chromosome modifications. Genomic differentiation would be more likely due to genetic divergence than to large structural genomic divergence, associated with an increase/decrease of repeated sequences with possibly transposable elements and rDNA gene families.
Some considerations can be made with respect to the timeframe of the chromosomal changes in the studied species. According to a dated phylogenetic tree of Solanaceae, S. vespertilio originated from a lineage that split and gave rise to S. sandwicense about 5.41 million years ago (Särkinen et al., 2013). This is during the same time period as when the cytogenetic differences observed in our work allegedly occurred (i.e. increase in heterochromatin in S. sandwicense). That amount of time is comparable to the age of the Acanthophora clade (4.71 mya), yet in this group the cytogenetic changes observed are comparatively more significant, involving reduction of chromosome length, percentage of heterochromatin and chromosome asymmetry and duplication of a 5S site (Chiarini et al., 2014).
Chromosome stasis in S. nelsonii, S. sandwicense and S. vespertilio is relative. Despite that each species demonstrated a conserved, basic chromosome number well associated to the Solanum genus, and karyotype formulae are not markedly different from other known "spiny solanums", we present evidence of cryptic, cumulative sequence changes. Thus, in these Solanum species studied, like other groups within the genus (Bernardello & Anderson, 1990; Bernardello et al., 1994; Chiarini et al., 2014), speciation has not been associated with large, obvious chromosome rearrangements nor with polyploidy.
Acknowledgements
Authors wish to acknowledge the assistance of the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) Agencia FONCyT and SECyT (Universidad Nacional de Córdoba, Argentina), for financial support and facilities used in this investigation.
Bibliography
1. ACOSTA, M.C., G. BERNARDELLO, M. GUERRA & E.A. MOSCONE. 2005. Karyotype analysis in several South American species of Solanum and Lycianthes rantonnei (Solanaceae). Taxon 54: 713–723. [ Links ]
2. ACOSTA, M. C., M. GUERRA & E. A. MOSCONE. 2012. Karyological relationships among some South American species of Solanum (Solanaceae) based on fluorochrome banding and nuclear DNA amount. Plant Syst. Evol. 298: 1547–1556. [ Links ]
3. ANDERSON, G.J., G. BERNARDELLO, L. BOHS, T. WEESE & A. SANTOS-GUERRA. 2006. Phylogeny and biogeography of the Canarian Solanum vespertilio and S. lidii (Solanaceae). Anales Jard. Bot. Madrid 63:159–167. [ Links ]
4. BERNARDELLO, L.M. & G.J. ANDERSON. 1990. Karyotypic studies in Solanum section Basarthrum (Solanaceae). Am. J. Bot. 77: 420–431. [ Links ]
5. BERNARDELLO, L.M., C.B. HEISER & M. PIAZZANO. 1994. Karyotypic studies in Solanum section Lasiocarpa (Solanaceae). Am. J. Bot 81: 95–103. [ Links ]
6. BOHS, L. 2005. Major clades in Solanum based on ndhF sequence data. In: KEATING, R. C., V. C. HOLLOWELL & T. B. CROAT (eds.), A Festschrift for William D'Arcy. Monogr. Syst. Bot. Missouri Bot. Gard. 104: 27–50. Missouri Botanical Garden Press, St. Louis, Missouri. [ Links ]
7. BRAMWELL, D. & J. CAUJAPÉ-CASTELLS (eds.) 2011. The biology of island floras. Cambridge University Press, Cambridge. [ Links ]
8. BRAMWELL, D. 2006. Medicinal plants of the Canary Islands. Medicinal Plants Conservation 12: 36-40. [ Links ]
9. CARLQUIST, S. 1974. Island biology. Columbia University Press, New York, USA. [ Links ]
10. CARR, G.D. 1985. Additional chromosome numbers of Hawaiian flowering plants. Pac. Sci. 39: 302–306. [ Links ]
11. CARR, G.D. 1998. Chromosome evolution and speciation in Hawaiian flowering plants. In: STUESSY, T.F. & M. ONO (eds.), Evolution and speciation of island plants 5-47. Cambridge University Press, Cambridge. [ Links ]
12. CHIARINI, F. & G. BERNARDELLO. 2006. Karyotypic studies in South American species of Solanum subgen. Leptostemonum (Solanaceae). Plant Biol. 8: 486–493. [ Links ]
13. CHIARINI, F., G. BERNARDELLO, G. ANDERSON & A. SANTOS GUERRA. 2006. Chromosomal differentiation of Solanum vespertilio and S. lidii (Solanaceae), rare, endemic species of the Canary Islands (Spain). Caryologia 59: 277-283. [ Links ]
14. CHIARINI, F., F.F. SANTIÑAQUE, J.D. URDAMPILLETA & M.L. LAS PEÑAS. 2014. Genome size and karyotype diversity in Solanum sect. Acanthophora (Solanaceae). Plant Syst. Evol. 300: 113-125. [ Links ]
15. DOVER, G.A. & R.B. FLAVELL. 1984. Molecular co-evolution: DNA divergence and the maintenance of function. Cell 38: 622–623. [ Links ]
16. GARCIA, S., T. GARNATJE, E.D. MC ARTHUR, J. PELLICER, S. SILJAK-YAKOVLEV & J. VALLÈS. 2009. Ribosomal DNA and genome size changes during polyploid and hybrid formation in the North American endemic sagebrushes (Artemisia, Asteraceae). Genome 52: 1012–1024. [ Links ]
17. GARCIA, S., F. GÁLVEZ, A. GRAS, T. GARNATJE & A. KOVARIK. 2013. Plant rDNA database. Release 2.0, including new features. http://www.plantrdnadatabase.com/ [accessed 04 June 2014]. [ Links ]
18. GERLACH, W.L. & J.R. BEDBROOK. 1979. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 7: 1869-1885. [ Links ]
19. GIVNISH, T.J., K.C. MILLAM, A.R. MAST, T.B. PATTERSON, T.J. THEIM, A.L. HIPP, J.M. HENSS, J.F. SMITH, K.R. WOOD & K.J. SYTSMA. 2009. Origin, adaptive radiation and diversification of the Hawaiian lobeliads (Asterales: Campanulaceae). Proc. Roy. Soc. B-Biol. Sci. 276: 407–416. [ Links ]
20. GUERRA, M. 1983. O uso do corante Giemsa na citogenetica vegetal: comparação entre a coloração simples e o bandeamento C. Ciência e Cultura 35: 190-193. [ Links ]
21. GUERRA, M. 2000. Patterns of heterochromatin distribution in plant chromosomes. Gen. Mol. Biol. 23: 1029–1041. [ Links ]
22. HUNZIKER, A.T. 2001. Genera Solanacearum. The genera of Solanaceae illustrated, arranged according to a new system. A.R.G. Gantner Verlag K.-G, Ruggell. [ Links ]
23. KITAMURA, S., M. INOUE, N. SHIKAZONO & A. TANAKA. 2001. Relationships among Nicotiana species revealed by the 5S rDNA spacer sequence and fluorescence in situ hybridization. Theor. Appl. Gen. 103: 678-686. [ Links ]
24. LEVAN, A., L. FREDGA & A. SANDBERG. 1964. Nomenclature for centromeric position on chromosomes. Hereditas 52: 201–220. [ Links ]
25. LEVIN, R.A., N.R. MYERS & L. BOHS. 2006. Phylogenetics relationships among the "Spiny Solanums" (Solanum subgenus Leptostemonum, Solanaceae). Am. J. Bot. 93: 157-169. [ Links ]
26. MALINSKA, H., J.A.TATE, R. MATYASEK, A.R. LEITCH, D.E. SOLTIS, P.S. SOLTIS & A. KOVARIK. 2010. Similar patterns of rDNA evolution in synthetic and recently formed natural populations of Tragopogon (Asteraceae) allotetraploids. BMC Evol. Biol. 10: 291.doi: 10.1186/1471-2148-10-291 [ Links ]
27. MELO, C.A.F., M.I.G. MARTINS, M.B.M. OLIVEIRA, A.M. BENKO-ISEPPON & R. CARVALHO. 2011. Karyotype analysis for diploid and polyploid species of the Solanum L. Plant Syst. Evol. 293: 227–235. [ Links ]
28. MOSCONE, E.A., M. LAMBROU & F. EHRENDORFER. 1996. Fluorescent chromosome banding in the cultivated species of Capsicum (Solanaceae). Plant Syst. Evol. 202: 37–63. [ Links ]
29. MOSCONE, E.A., M. SCALDAFERRO, M. GRABIELE, N. CECCHINI, Y. SANCHEZ GARCÍA, R. JARRET, J. DAVIÑA, D. DUCASSE, G. BARBOZA, F. EHERENDORFER. 2007. The evolution of chili peppers (Capsicum - Solanaceae): a cytogenetic perspective. Acta Hort. 745: 137-169. [ Links ]
30. MOYETTA, N.R., L.B. STIEFKENS & G. BERNARDELLO. 2013. Karyotypes of South American species of the Morelloid and Dulcamaroid clades (Solanum, Solanaceae). Caryologia 66: 333- 345. [ Links ]
31. OLMSTEAD, R.G., L. BOHS, M. HALA ABDEL, E. SANTIAGO-VALENTIN, V.F. GARCIA & S.M. COLLIER. 2008. A molecular phylogeny of the Solanaceae. Taxon 57:1159–1181. [ Links ]
32. PROHENS, J., G. ANDERSON, F. HERRAIZ, G. BERNARDELLO, A. SANTOS-GUERRA, D. CRAWFORD & F. NUEZ. 2007. Genetic diversity and conservation of two endangered eggplant relatives (Solanum vespertilio Aiton and Solanum lidii Sunding) endemic to the Canary Islands. Gen. Res. Crop Evol. 54: 451-464. [ Links ]
33. REGO, L.N.A.A., C.R.M. DA SILVA, J.M.D. TOREZAN, M.L. GAETA & A.L.L. VANZELA. 2009. Cytotaxonomical study in Brazilian species of Solanum, Lycianthes and Vassobia (Solanaceae). Plant Syst. Evol. 279: 93–102. [ Links ]
34. ROMERO ZARCO, C. 1986. A new method for estimating karyotype asymmetry. Taxon 35: 526- 530. [ Links ]
35. SANDERS, R.W., T.F. STUESSY & R. RODRIGUEZ. 1983. Chromosome numbers from the flora of the Juan Fernandez Islands. Am. J. Bot. 70: 799–810. [ Links ]
34. SÄRKINEN, T., L. BOHS, R.G. OLMSTEAD & S. KNAPP. 2013. A phylogenetic framework for evolutionary study of the nightshades (Solanaceae): a dated1000-tip tree. BMC Evol. Biol. 13:214. doi:10.1186/1471-2148-13-214 [ Links ]
35. SCALDAFERRO, M.A., M. GRABIELE & E.A. MOSCONE. 2013. Heterochromatin type, amount and distribution in wild species of chili peppers (Capsicum, Solanaceae). Gen. Res. Crop Evol. 60: 693-709. [ Links ]
36. SCHWARZACHER, T. & P. HESLOP-HARRISON. 2000. Practical in situ hybridization. Bios Scientific Publishers Limited, Oxford. [ Links ]
37. SCHWEIZER, D. 1976. Reverse fluorescent chromosome banding with chromomycin and DAPI. Chromosoma (Berlin) 58: 307-324. [ Links ]
38. SCHWEIZER, D. & P. AMBROS. 1994. Chromosome banding. In: GOSDEN, J.R. (ed.), Methods in molecular biology, Chromosome analysis protocols, 97-112. Humana Press, Totowa. [ Links ]
39. SCHWEIZER, D. & J. LOIDL. 1987. A model for heterochromatin dispersion and the evolution of C-band patterns. In: STAHL, A. et al. (eds.), Chromosomes Today, pp. 61–74. Allen & Unwin, London. [ Links ]
40. STUESSY, T.F. & D.J. CRAWFORD. 1998. Chromosomal stasis during speciation in angiosperms of oceanic islands. In: STUESSY, T.F. & M. ONO (eds.), Evolution and speciation of island plants 307-332. Cambridge University Press, Cambridge. [ Links ]
41. STUESSY, T.F. & M. ONO (eds.) 1998. Evolution and speciation of island plants. Cambridge University Press, Cambridge. [ Links ]
42. VITOUSEK, P.M., L.L. LOOPE & H. ADSERSEN (eds.) 1995. Islands: Biological Diversity and Ecosystem Function. Ecological Studies vol. 115, Springer, Heidelberg. [ Links ]
43. VORONTSOVA, M., S. STERN, L. BOHS & S. KNAPP. 2013. African spiny Solanum (subgenus Leptostemonum, Solanaceae): a thorny phylogenetic tangle. Bot. J. Linn. Soc. 173: 176–193. [ Links ]
44. WAGNER, W.L. & V. FUNK (eds.). 1995. Hawaiian Biogeography: Evolution on a Hot-Spot Archipelago. Smithsonian Institution Press, Washington. [ Links ]
45. WAGNER, W.L., D.R. HERBST & D.H. LORENCE. 2005. Flora of the Hawaiian Islands website. http://botany.si.edu/pacificislandbiodiversity/hawaiianflora/index.htm [accessed 18 Dec 2013] [ Links ]
46. WEESE, T. & L. BOHS. 2007. A Three-Gene phylogeny of the genus Solanum (Solanaceae). Syst. Bot. 32: 445–463 [ Links ]
47. WEISS, H., B-Y. SUN, T.F. STUESSY, C.H. KIM, H. KATO & M. WAKABASHI. 2002. Karyology of plant species endemic to Ullung Island (Korea) and selected relatives in peninsular Korea and Japan. Bot. J. Linn. Soc. 138: 93-105. [ Links ]
48. WHALEN, M.D. 1984. Conspectus of species groups in Solanum subgenus Leptostemonum. Gentes Herb. 12: 179-282. [ Links ]
Recibido el 6 de noviembre de 2015,
aceptado el 10 de febrero de 2016.














