Introduction
One of the taxonomic approaches most intensively explored in the last century was cytotaxonomy [summarized by Stebbins (1971)]. The rationale behind this approach is that the karyotype allows a “macroscopic” view of the genetic material of a species, with some particular details of individuals or populations and reveals many characters common to its genus or tribe. In contrast with various other morphological descriptors, karyotype changes, such as polyploidy and chromosome inversions or translocations, may work as reproductive barriers among populations or species, playing therefore an important role in the speciation process (Levin, 2002). Moreover, the karyotype is the only morphological trait that is not affected by gene expression, environmental conditions, age, developmental phase, etc (Guerra, 2012). In general, using appropriate techniques, karyotype similarity among species of a genus is an indicative of their phylogenetic proximity.
Karyotype description was initially restricted to chromosome number, size and morphology (Stebbins, 1971), but recent advances on molecular cytogenetics expanded the number of chromosomal marks and allowed a more detailed karyotype description (Jiang, 2019). For example, the chromosomes of all species of Citrus and related genera are very similar in number, size and morphology but they differ widely in the number and distribution of heterochromatic bands and 5S and 35S rDNA sites (reviewed by Guerra, 2009). Likewise, the assumption that the families Juncaceae and Cyperaceae had holocentric chromosomes as one of their synapomorphies (Judd et al., 2016) was overturned by careful analyses of the chromosome morphology and centromere immunostaining of some Juncus species, revealing that in Juncaceae, at least, holocentricity is rather a particularity of a few genera (Guerra et al., 2019).
Structural chromosome differentiation is most commonly revealed by identification of heterochromatic bands and sites of highly conserved DNA sequences, as the telomeric DNA and 5S and 35S rRNA genes. These marks are composed by repetitive DNA sequences densely concentrated into a few blocks and located preferentially at the centromeric and terminal chromosome regions or near the nucleolus organizer region (Guerra, 2000; Roa & Guerra, 2015; Samoluk et al., 2017). Within certain limits, the amount of heterochromatin is not critical to genome function but in a few species it is enormously expanded, forming large heterochromatic blocks, as in Trithrinax campestris and Capsicum species (Gaiero et al., 2012; Grabiele et al., 2018), or numerous small bands, as in Cuscuta monogyna (Ibiapino et al., 2020). The tandemly repeated nature of rDNA and telomeric DNA sites allows a considerable variation in number and size of these sites, contributing to a better karyotype characterization of species or populations (Pedrosa-Harand et al., 2006; Robledo & Seijo, 2010; Rosato et al., 2017, 2018; Silvestri et al., 2020). Currently, chromosome staining with the fluorochromes chromomycin A3 (CMA) and 4’,6-diamidino-2- phenylindole (DAPI) is the most used method to differentiate GC-rich and AT-rich heterochromatic bands, respectively (Guerra, 2000). Other more specific chromosome regions are revealed by fluorescence in situ hybridization (FISH), a well stablished method to localize any type of DNA sequence along the chromosomes (Jiang, 2019). The analysis of CMA/DAPI bands and rDNA sites, or other DNA sequences revealed by FISH, into a phylogenetic context has ensured an enormous progress in cytotaxonomical analyses (e.g., Chalup et al, 2015; Silvestri et al., 2020; Ribeiro et al., 2020). However, cy to molecular methods are more time-consuming and have been used in a still limited number of cytotaxonomical works.
Zephyranthes brachyandra (Baker) Backer (Amaryllidaceae) is an ornamental bulbous plant native of the Neotropics, from southern South America to Mexico and southwest USA, including southern Brazil and West Indies, sometimes referred to as Habranthus brachyandrus (Baker) Sealy (Daviña, 2001; Daviña & Honfi, 2018; García et al. 2019). Phylogenetically, the former genera Zephyranthes and Habranthus are now recognized as subgenera of Zephyranthes s.l. (García et al. 2019). The two subgenera share similar chromosome size and morphology, large variation in chromosome numbers, mainly with x = 6, several ploidy levels, and some species with B chromosomes (Naranjo, 1974; Felix et al., 2008, 2011a; Daviña, 2001; Daviña & Honfi, 2018).
Cytologically, Z. brachyandra is a tetraploid with 2n = 24, characterized by many DAPI+ and CMA+ heterochromatic bands located in the interstitial- terminal region of most chromosome arms (Felix et al., 2011b). The individual analyzed by Felix et al. (2011b) presented 2n = 24 plus one B chromosome, whereas the five bulbs examined by Davina (2001) exhibited always 2n = 24. B chromosomes are extra chromosome usually smaller than the regular chromosomes of the species (A chromosomes), mostly depleted of genes, and present only in some individuals of the species (see, e.g, Marques et al, 2013; Vanzela et al, 2017). They may occasionally be accumulated in some individuals affecting their DNA content and some other phenotype characters (Huang et al, 2016).
In the present work, we conducted a detailed karyotype analysis of three individuals of Z. brachyandra collected in the field, aiming to investigate: a, the stability of the many CMA+ and DAPI+ bands reported previously for a single individual (Felix et al. 2011b); b, the distribution of 5S rDNA, 35S rDNA and telomeric DNA sites; c, the occurrence of B chromosomes. We also evaluated karyotype similarity in chromosome number, size and morphology, banding patterns and rDNA sites of Z. brachiandra with other species of the genus, in order to contribute to the cytotaxonomy of Zephyranthes.
Material and Methods
Plants material
Three bulbs of Z. brachyandra were collected in San Javier (27°52'17.9”- 55°07'41.7”), Misiones, Argentina, and cultivated in the Experimental Garden of the Department of Botany, Federal University of Pernambuco, Recife, Brazil. A voucher was deposited in the herbarium Prof. Jayme Coelho de Moraes (Federal University of Paraiba, Brazil, voucher EAN 29460).
Slides preparation
For mitotic analyses, young root tips were pretreated with colchicine 0.2% at 10 °C for 24 h, fixed in ethanol-acetic acid (3:1, v/v) for 2 h at room temperature, then stored at -20 °C until the moment of use. The fixed meristems were washed twice in destilled water, digested in a 2% cellulase (Onozuka)/ 20% pectinase (Sigma) solution at 37 °C for one hour, and macerated in a drop of 45% acetic acid. The coverslips were removed in liquid nitrogen and the slides were dried in the air. The preparations were mounted and stained with a 1 pg/mL DAPI/glycerol (1:1) solution, in order to select the best slides. Subsequently, they were destained in ethanol/acetic acid (3:1), air dried, and aged for three days at room temperature.
Chromosome staining with CMA/DAPI and FISH
Chromosome double staining with the fluorochromes CMA (Sigma) and DAPI (Sigma) was performed as described by Vaio et al. (2018). Aged slides were stained with CMA (0.1 mg/mL) for 1 h, counterstained with DAPI (2 pg/mL), mounted with glycerol-Mcllvaine buffer pH 7.0 (1:1) and aged again for three days. After image capture of the best metaphases the chromosomes were destained and in situ hybridized according to Pedrosa et al. (2002). The probes used for 5S and 35S rDNA sites were, respectively, D2 from Lotus japonicus (Regel) K. Larsen and a clone from Triticum aestivum L. (plasmid Pta71). The probes were direct and indirect labeled with Cy3 dUTP (GE Healthcare) (5S rDNA probe) and digoxigenin-11-dUTP (Roche) (35S rDNA probe) respectively, both by nick translation (Invitrogen). The hybridization mixture contained 50% formamide (v/v), 10% dextran sulfate (w/v), 2x SSC, and 5-10 ng/pL of rDNA probe, accomplishing a final stringency of ~76%. The 35S rDNA probe was detected with sheep anti- digoxigenin-FITC (Roche).
For detection of telomeric sites, a synthetic TTTAGGG oligoprobe labeled with Cy3 by Macrogen Inc. was in situ hybridized according to the protocol of Cuadrado et al. (2010), slightly modified. Briefly, 10 pL of the probe solution (8 ng/pL of the oligoprobe diluted in 2x SSC) were applied to each slide for 2 h at room temperature. Afterwards, the slides were washed in 4x SSC/0.2% Tween20 for 10 min. All preparations were counterstained and mounted with 2 pg/ mL DAPI in Vectashield (Vector). Images of the best cells were captured with a Leica DM5500B fluorescence microscope and later processed with Adobe Photoshop CC 2020 for brightness, contrast, and sharpness.
Chromosome measurements
In order to characterize chromosome size and morphology, chromosomes of the three best metaphases for each karyotype were measured using the software DRAWID version 0.26 (Kirov et al, 2017). The chromosome arm ratio [r = length of long arm (l)/short arm (s)] was used to define chromosomes as metacentric (r = 1.0 1.49), submetacentric (r = 1.50-2.99), acrocentric (AR > 3.0) or telocentric, according to Guerra (1986). Chromosome pairs were ordered from I to XII by decreasing size of the short arm.
Results
The plants collected in Argentina grew very well and flowered during the summer in Recife (Fig. 1), but only produced sterile seeds. They revealed a high rate of asexual reproduction by lateral bulb production, providing plenty of material for investigation. About 15 metaphases of each individual were analyzed for chromosome number. Two individuals analyzed presented 2n = 24 and a third presented 2n = 24 plus a B chromosome. The three best metaphases of each plant were selected for a careful comparison of CMA/DAPI bands and rDNA sites. Two karyograms were mounted for each individual and a single ideogram was constructed summarizing the average sizes for each chromosome pair and heterochromatic bands and rDNA sites common to all of them (Fig. 2). The individuals without B chromosomes were heteromorphic for a chromosome pair formed by a meta- and a submetacentric chromosome and were karyotypically identical regarding all other chromosome characteristics, suggesting that they were clones from a single individual. Therefore, they will be referred to herein as a single karyotype. Excluding the B chromosome, chromosome size varied from 14.59 to 9.28 pm and the average haploid complement size was 146.75 pm (Fig. 2 and Table 1). The chromosome morphology in the two karyotypes was very similar, except for the arm ratio of chromosome pair X, which was 1.24 for the karyotype without B and 1.80 for the karyotype with B. This difference changed its chromosome morphology from metacentric (AR = 1.00-1.49) to submetacentric (AR = 1.50-2.99) (Guerra, 1986) and the karyotype formula from 8M + 4SM for the individual with 2n = 24 to 9M + 3SM for the individual with 2n = 24 + 1B (the chromosome B was not included in the karyotype formula). The average arm ratio for pair X considering both karyotypes was 1.52 (Fig.2). The remaining chromosomes were metacentrics (pairs I to IX) or submetacentrics (pairs XI and XII). The most outstanding karyotype feature of Z. brachyandra was the large amount of DAPI+ heterochromatin. There were large DAPI+ bands on the short arm of 10 chromosomes and one to four small bands in the interstitial/terminal region of the remaining chromosome arms (Fig. 3A). The submetacentric pairs XI and XII presented similar size and morphology, differing mainly in the number of DAPI+ bands on the long arm: two on pair XI and three on pair XII. CMA+ bands were identified only in the short arms of the smaller submetacentrics (Fig. 3A’) and in the short arm of pair VIII (not always visible). Besides these general features, the two karyotypes observed here had several small structural differences.
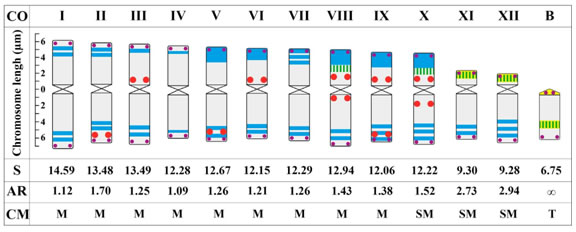
Fig. 2: Idiogram of Z. brachyandra, including all characters observed in both karyotypes plus the B chromosome. CO, chromosome order; S, chromosome size in μm; AR, arm ratio; CM, chromosome morphology. The scale on the left side indicates the size of the chromosome arms. Blue = DAPI+ band; yellow = CMA+ band; yellow with green bars = 35S rDNA sites co-localized with CMA+ bands; red dots = 5S rDNA sites; very small purple dots = TTTAGGG sites.
The number and position of the 5S and 35S rDNA loci complemented the identification of each chromosome pair and were different in the two karyotypes. The karyotype with the heteromorphic pair had 19 sites of 5S and six sites of 35S rDNA. The 5S rDNA sites were located on the interstitial (12 sites) or terminal (7 sites) regions of the large metacentric and submetacentric chromosomes and a single site on one homologue of pair XI (Fig 3A’’). The five small submetacentrics had a 35 S rDNA site on the short arm co-localized with the CMA+ band. One chromosome of pair VIII also displayed a small 35S rDNA near the large DAPI+ band, but it was not co-localized with a CMA+ band. The karyograms based on Fig. 3A-A’’ placed the extra small submetacentric provisionally as pair IV (Fig. 3B), since one homologue of pair IV was absent. However, it was almost identical to pair XI in size and morphology as well as in banding patterns, with two weakly differentiated DAPI+ bands on the long arm and a CMA+ band on the short one. At least six other chromosome pairs were heteromorphic for one or more bands or sites (asterisks in figure 3B).
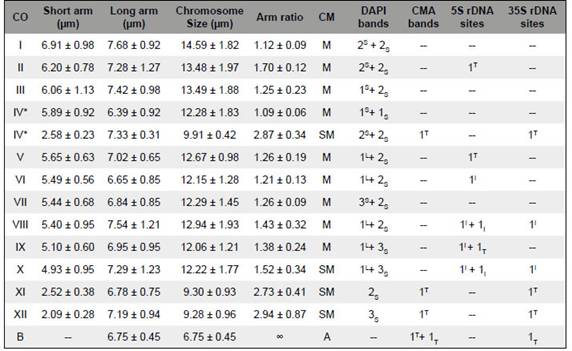
Table 1: Chromosome data of Zephyranthes brachyandra karyotype, including chromosome order (CO, larger to smaller short arm), short and long arm sizes, chromosome size, arm ratio, chromosome morphology (CM), number and position of DAPI+ bands and CMA+ bands, and number and position of 5S and 35S rDNA sites. DAPI+ bands were classified as large (L) or small (S), CMA+ bands as terminal (T), and rDNA sites as terminal (T) or interstitial (i). Chromosomal marks on the short arms are superscript and those on the long arms are subscript.
The karyotype with a B chromosome exhibited a similar pattern of CMA/DAPI bands and rDNA sites (Fig. 4A-A’). It had 19 sites of 5S rDNA, including an extra signal on the short arm of both homologues of pair V and a heteromorphic site on pair X, and 12 sites of 35S rDNA plus one in the B chromosome. Noteworthy, the additional 35S rDNA sites, located on pairs VIII and X, were not clearly detected as CMA+ bands. The 35S rDNA site on pair X was very close to the large DAPI+ band but they were not colocalized (see insets in Fig. 4A and respective chromosomes in Fig. 4B).
The TTTAGGG probe labelled the telomeric regions of all chromosomes of both karyotypes and no ectopic site was found (Fig. 5). No short arm was visible on the B chromosome, suggesting that it was a telocentric. It was shorter (6.75 pm) than the submetacentrics (Fig. 2), had no DAPI+ bands, and presented two CMA+ bands: one interstitial and one very small on its narrower end (inset in Fig. 4A’). The latter could be the centromere itself or a very small short arm, whereas the interstitial band coincided with a 35S rDNA site.
Discussion
The two karyotypes of Z. brachyandra analysed here differed mainly in the occurrence of a heteromorphic chromosome pair in one of them and a B chromosome in the other. Regarding chromosome sizes and haploid karyotype formula, our data (14.59 to 9.28 pm and 8M + 4SM or 9M + 3SM) differ in part from that reported by Davina (2001) (10.89 to 6.14 pm and 8M + 4SM). The difference in chromosome size may be due to the distinct anti-mitotic pre-treatment and the condensation status of selected metaphases (Guerra, 2012), whereas the karyotype formula 8M + 4SM was the same found here for the karyotype without B. Therefore, in spite of the small sample size, our data revealed karyotype instability, which was not detected in previous studies.
The two karyotypes displayed a similar number of CMA/DAPI bands and differed slightly in the number of the 5S and 35S rDNA sites. The complex DAPI+ banding pattern of Z. brachyandra wassimilar to that reported by Felix et al. (201 lb), while the number and position of CMA+ bands reported by these authors were quite distinct from the present ones. They found eight or ten bright CMA+ bands (excepting the B chromosome), only two of them on small submetacentric chromosomes, whereas in our sample there were only four or six CMA+ bands (excepting the B and the extra submetacentric chromosome). It is possible that some CMA+ bands in our sample had not been detected because they were too small. Actually, the 35S rDNA sites of pairs VIII and X were expected to be CMA+, since they are usually co-localized with CMA+ bands (e.g., Moraes et al., 2007; Gaiero et al, 2012; Silva et al., 2019). However, small rDNA sites, like those, are sometimes not detected as CMA+ bands (e.g., Vaio et al, 2018). Variation in size and number of heterochromatic bands and rDNA sites depends on the number of repetitive sequences per locus, which may be caused by recombination events with unequal exchanges (Lower et al, 2018). However, the position of some interstitial CMA+ bands reported by Felix et al. (2011b) did not coincide with position of the 35S rDNA sites found in our sample, suggesting that if they correspond to 35S rDNA sites, they are not the same reported here.
B chromosomes probably arise from regular A chromosomes and can alter their original DNA composition by rapidly accumulating or losing several DNA sequences (Marques et al, 2013). In Z. brachyandra the only B chromosome observed was a telocentric chromosome similar in size and morphology to that reported by Felix et al. (2011a,b), but without the two interstitial DAPI+ bands observed by those authors. Both Bs had a small terminal CMA+ band, although only one reported here had an extra interstitial CMA+ band co-localized with a 35S rDNA site. Polymorphisms on B chromosomes are more common than in the regular chromosomes, probably due to the dispensable nature of Bs (Marques et al, 2013; Vanzela et al, 2017).
At first sight, the extra submetacentric chromosome found in a karyotype with the normal chromosome number seemed to be a partial deletion of the short arm of a metacentric chromosome. Heteromorphism for large chromosome segments are rare but have already been clearly documented for several plant species, especially those with large chromosomes, as Crocus cancellatus (Brighton, 1976) and Alloe rabaiensis (Bradham, 1983). However, the similarity of the extra submetacentric with chromosome pair XI in size, morphology, and CMA/DAPI bands, as well as the absence of one homologue of pair IV, suggest that this karyotype had a trisomy for pair XI combined and a monosomy for pair IV. Such monotrisomic aneuploids are more commonly found among chromosomically engineered crop plants, as wheat and barley, and most of them are compensating aneuploids involving homeologous chromosomes (Singh, 2003). Differently, in Z. brachyandra the monotrisomy involved two quite different chromosomes, although the plant phenotype was identical to the normal plants. In this case, the normal appearance of the individual is most probably due to the buffer effect of polyploidy, in which a deletion or a duplication of an entire chromosome of a tetraploid genome could not be sufficient to alter the phenotype (Deng et al., 2018). The occurrence of aneuploid plants is more common in meiotically instable polyploids (Mandáková & Lysak, 2018), while Z. brachyandra has a regular meiosis (Daviña, 2001; Daviña & Honfi, 2018). Monotrisomy in wild plants has rarely been reported, but it is probably underestimated because its detection demands a clear distinction of the chromosomes involved, as demonstrated in Tragopogon miscellus (Chester et al., 2012) and Nothoschordum bonariense (Souza et al, 2019). Trisomy as well as B chromosomes have also been reported in some other species of Zephyranthes (see e.g., Felix et al., 2008, 2011a,b).
Despite the polymorphisms observed here, Z. brachyandra is karyotypically very well defined, due to its many chromosome marks. The only other Zephyranthes species where rDNA sites have been investigated is Z. robusta, a diploid with 2n = 12 (10M + 2SM) and similar chromosome sizes. The molecular karyotype of Z. robusta is also similar to that expected for a hypothetical diploid ancestor of Z. brachyanda, with two 35S rDNA sites co localized with CMA+ bands on submetacentric short arms, ten 5S rDNA sites, some interstitial and subterminal DAPI+ bands and a few other heterochromatic blocks detected by C-banding (Barros e Silva & Guerra, 2010; Felix et al, 2011b; A.E. Barros e Silva, unpublished results). This similarity points to a possible autopolyploid origin for Z. brachiandra, in spite of its regular meiotic pairing (Daviña and Honfi, 2018), which
is usually associated with allopolyploidy. Regular bivalent formation has also been reported for other autopolyploids, as Arabidopsis arenosa (Carvalho et al., 2010), and autopolyploidy may be not as rare as supposed earlier (Soltis et al., 2007). The assumption of an autopolyploid origin for Z. brachiandra could also explain the many duplicated patterns of chromosome banding and rDNA site distribution observed here, some of them blurred by the intense heteromorphism characteristic of heterochromatic bands and rDNA sites (Chalup et al., 2015; Silvestri et al., 2020; Ribeiro et al., 2021). Five other species of Zephyranthes analysed by Felix et al. (2011b) revealed a much smaller and variable number of CMA+ bands and no DAPI+ bands, suggesting that a cytomolecular investigation of other Zephyranthes species would be very helpful to understand the cytotaxonomy and chromosomal evolution of this group.
Authors Contribution
THN conducted the FISH experiments, analyzed the data, constructed the figures and tables and wrote the paper. RSG conducted FISH experiments and helped to write the paper. MB collected the material, conducted FISH experiments and helped to write the paper. GS collected the material and helped to write the paper. MG discussed the results, provided resources for the FISH experiments, designed this research and wrote the paper. All authors read and approved the final version of the paper.












 uBio
uBio 


