Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
Phyton (Buenos Aires)
versión On-line ISSN 1851-5657
Phyton (B. Aires) v.77 Vicente López ene./dic. 2008
ARTÍCULOS ORIGINALES
Quantitative measures of leaf epidermal cells as a taxonomic and phylogenetic tool for the identification of Stanhopea species (Orchidaceae) (With 3 Tables & 2 Figures)
Mediciones cuantitativas en las células epidérmicas de la hoja como herramienta taxonómica y filogenética para la identificación de especies de Stanhopea (Orchidaceae) (Con 3 Tablas y 2 Figuras)
Foroughbakhch R, RJ Ferry Sr, JL Hernández-Piñero, MA Alvarado-Vázquez, A Rocha-Estrada
Facultad de Ciencias Biológicas, UANL, Depto. de Botánica, A.P. F-2, 66451 San Nicolás de los Garza, N.L, México. Phone: 52-81-8114 3465. Address correspondence to: Rahim Foroughbakhch P., e-mail: rahim.f@gmail.com, rahimforo@hotmail.com
Recibido/ Received 28.06.2007. Aceptado/Accepted 26.02.2008.
Abstract. Orchids of the genus Stanhopea are currently identified only by their floral structure characteristics. A statistical analysis of a significant number of species of this genus disclosed that measurements of adaxial and abaxial epidermal cell surface areas can be correlated with specific recognized species by a leaf printing method. This allows an objectively either positive or nearly positive confirmation of the identity of a species in the absence of flowers and without damage to plants. When ordering the mean values obtained for these surface areas in each species in a decreasing order, a correlation was observed in a hierarchical way that went from primitive to more advanced floral forms. This reflects the evolutionary radiation of the genus. It is established that in Stanhopea, the presence of large leaf epidermal cells on species from South America represents a primitive evolutionary condition that became to smaller cells in evolutionarily more recent individuals as the genus radiated towards Mexico.
Key Words: Stanhopea; Quantitative taxonomy; Morphometry; Leaf printing; Orchidaceae.
Resumen. Las orquídeas del género Stanhopea son identificadas actualmente solamente por las características estructurales de la flor. El análisis estadístico de un número significativo de especies de este género reveló que las áreas promedio de las células epidérmicas adaxiales y abaxiales se pueden correlacionar con especies específicamente reconocidas utilizando un método de impresión de la hoja. Esto permitiría una confirmación positiva o casi positiva de la identidad de una especie de modo objetivo, en ausencia de flores y sin daño a las plantas. Al ordenar los valores obtenidos para dichas áreas de las células epidérmicas en cada especie en un orden descendente, se observó una correlación de modo jerárquico que va de las formas florales primitivas a las formas más avanzadas. Esto reflejó una radiación evolutiva del género. Se establece que en Stanhopea, las grandes células epidérmicas presentes en las hojas de especies de Sudamérica representan una condición evolutiva primitiva que fue cambiando a células más pequeñas en individuos evolutivamente más recientes a medida que el género se expandió hacia México.
Palabras clave: Stanhopea; Taxonomía cuantitativa; Morfometría; Impresión de hoja; Orchidaceae.
INTRODUCTION
The collection, pressing and drying of plant samples, which are then analyzed for its classification and taxonomical identification, has been the classical botanical protocol to investigate and describe the flora of a particular region. If any plant sample could not be recognized as a known species, it was then considered as a new species. Classical taxonomy stresses the need of selecting and describing a "type" from a plant population, which is then deposited in a recognized herbarium, to validly describe a new species. This information might then be published in a high impact factor scientific Journal. However, because of the importance historically placed by Linnaeus on floral structures, many orchids have been described from just the floral structures, disregarding any other organ for taxonomical classification.
On the other hand, most new orchid species have been described by foreign botanists residing in remote localities from the plant's native area. They have mostly worked with floral specimens either dried or pressed or preserved in alcohol or some other fixative solution. For many years of modern orchid taxonomy, thousands of plants were collected from the New World tropical countries and shipped to greenhouses and taxonomists of Belgium, England, Germany, France and the USA. Although many plants survived, thousands of them were taken out from their habitats and either died in transit or during the establishment phase because of improper transporting and/or ignorance of their cultivation requirements.
In the 1960's, orchid conservationists in the United States and tropical countries achieved an international ecological awareness which resulted in the 1973 United States Federal Endangered Species Act. This was followed by an international agreement designed to preserve endangered species, prohibiting either the movement or export of any orchid specimen from its native habitat. However and despite these protecting and regulation laws, the illegal collection and traffic of orchid plants still continues.
The present situation is multifaceted. Orchid conservation requires species identification with techniques which must not be in conflict with existing laws and regulations. Also, identification based only on floral characteristics requires finding these relatively scarce plants when flowering. Precise knowledge of the species morphological and anatomical characteristics may allow a rapid and efficacious method of plant recognition. This would help to solve the above mentioned constraints.
The taxonomy of Stanhopea has been based solely on floral characters with only a note about vegetative structures (Arnold, 1928; Dodson & Frymire, 1961; Dodson, 1963, 1975). Furthermore, Curry et al. (1988) reported that the taxonomy of Stanhopea rests exclusively on flower morphological differences, which have been influenced by pollinators. This system of genus classification certainly has merit, but problems arise when either the boundary line of floral characters between close-related species is relatively indistinct or no flowers are present. Therefore, development of a system capable of reasonably and objectively correlate and identify a specimen against an already-described species is necessary. This method, by use of other than floral characteristics, would provide a useful identification tool at any time and place without regard to restrictions imposed by the flowering period.
Stanhopea was selected for this study due to certain attributes which make it a rigorous test genus. Flowers of Stanhopea do not last more than two or three days. It also exhibits few gross morphological differences at the vegetative developmental stage. Most of the literature only describes this aspect as "typical for the genus". If identifications are to be made using present classical methods, they must be done when plants are either flow-ering in their habitats or removed to a more convenient location to the botanist for observation when it does flower.
It may appear to be a relatively simple task to collect a plant, take care of it in a greenhouse, and then identify it when it flowers. However, the new habitat may not be appropriate to induce flowering because greenhouse conditions are far different from those in its natural habitat. This is in addition to the legal and regulatory problems related to the collection and transport of plant material.
Stanhopea is longitudinally widespread throughout the neotropical and tropical latitudes of the Western hemisphere. Because of this, it was hypothesized that an other-than-flower identification system might provide clues concerning intra-species relationships and the evolutionary radiation pattern of this genus. Specifically, it was hypothesized that identification of unknown species can be determined by comparing their mean leaf epidermal cell surface areas with those of known species.
Differences in morphometric characteristics of leaf epidermal cells and other structures (i.e., stomata) have been reported for different species of the genus Stanhopea (Ferry et al., 1997). An identification protocol will be described which (1) does not make changes to plants in the field, (2) requires no material to be taken from the habitat, (3) it is economical and easy to do, and (4) it is statistically supported rather than being a subjective description. Results using morphological differences on a great portion of the entire genus Stanhopea in this study, which may be intrinsic or due to geographical habitat changes, allowed to achieve a statistical method for the accurate identification of species in the absence of flowers.
MATERIALS AND METHODS
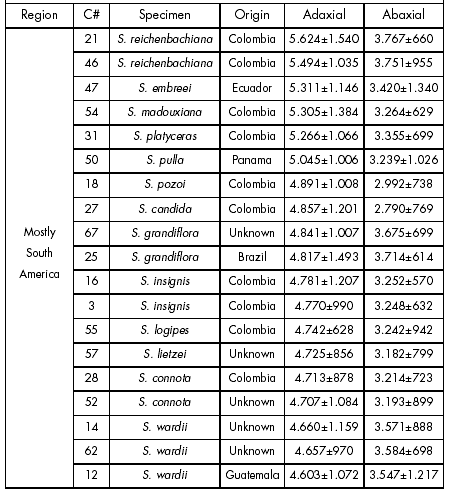
Biological material. Fifty four specimens of the genus Stanhopea were obtained which represented 34 species (Table 1). Although some species names are listed more than once in this table, no species or hybrid material represents the same clonal material. Among the samples, six Stanhopea specimens were received as unknown species (C03, 26, 33, 53, 77 and 83). Data obtained from those unknown species were statistically checked against similar data obtained from species confirmed by flowering. Subsequent flowering by the unknown species enabled to validate the statistically derived prediction.
Table 1. Summary of data obtained from the analyzed Stanhopea specimens. Data shown are epidermal cell surface areas (μ2). Each value is the mean ± 1 s.e. (n=200).
Tabla 1. Resumen de datos obtenidos de los especimenes de Stanhopea analizados. Los datos que se muestran son superficies celulares epidérmicas (μ2). Cada valor es el promedio± 1 e.e. (n=200).


Leaf-printing. The method used to obtain leaf-prints produces best results when the leaf surface areas are clean. Under greenhouse or field conditions, residues on the leaf surface areas must be eliminated by washing both adaxial and abaxial leaf surfaces with water, gently wiping them with a Teflon kitchen sponge, and allowing them to air-dry for a few minutes. If the plant had been exposed to an excessive amount of dust or residues, it may be useful to employ a very mild detergent, followed by rinsing in water and allowing the leaf to dry. Clean white Styrofoam "popcorn" was dissolved in xylol until the liquid was about the viscosity of warm syrup. This solution was applied to a clean leaf surface over an area of 2 x 5 cm. After allowing to air-dry for 2-3 minutes, a short strip of clear, transparent tape was applied evenly and firmly over the film with enough pressure as not to damage the leaf cells. The tape was peeled and gently passed onto a glass slide. A thin glass coverslip may be applied if permanent conservation of the slide is desired. This not only ensures that the leaf print is held flat but gives some protection to the transparent tape itself. Date, location and the study leaf surface area (either adaxial or abaxial), should be marked on the slide.
Photographic recording and cell measurements. Slides were placed under a microscope and photographed at a 80x magnification using a compound microscope with a trinocular head on which a Nikon HFM photo system was mounted. An indexing lens was inserted in the field lens assembly to print an index mark on each photomicrograph which enable to take accu-rate measurements directly from the print. A Reichert-Jung micrometer stage slide of 2 mm, divided into units of 0.01mm, was used to establish the lens correction factor for the magnification of each objective lens. Cell print photomicrographs were obtained randomly from the glass slides, utilizing a minimum of two photographs for each leaf surface area on each specimen. After dividing each photograph into 8 equal quadrants, 25 cells were randomly selected, and measurements of their length and width were individually recorded to give a closest approximation to the correct leaf cell surface area.
Statistical analysis. Individual specimen spreadsheets were computer-linked which provided a combined summary data sheet for each specimen; it also allowed specimen ranking according to their leaf epidermal cell areas, arranged in ascending or descending order. Data from individual adaxial and abaxial leaf epidermal cells of confirmed species were compared statistically. Entering individual data measurements into a statistical program (SAS ver. 6.12) provided a computer-generated analysis of variance readout (p= 0.05) for each specimen.
Data obtained from unknown Stanhopea samples were statistically compared (p= 0.05) with previously correlated data from confirmed species, and predictions were made on their identity. As mentioned above, validity of the predictions was checked out by identification of the samples after their flowering.
To double-check data accuracy, 10 samples were randomly selected and 200 cells were measured for each parameter, which was compared with the original samples (data not shown); an error margin less than 0.01% was used with this purpose. Four other randomly selected samples were measured for another individual without knowledge of the original data. Results of the 4 samples were less than 2% different from those of the original data.
RESULTS
Taxonomical differentiation. Sorting of the epidermal cell surface areas in a descending order showed the existence of gaps between most of the analyzed species (Table 1). Epidermal cell surface areas were significantly different (p<0.05) among species. The following results from selected examples will describe the process and efficacy of the method for species identification.
Four samples of S. tigrina were analyzed showing that none of the specimens differed significantly (p= 0.05). It was confirmed that these samples originated in Colombia, Guatemala and Mexico which demonstrates that this species does not vary significantly with latitude.
When the mean epidermal cell surface areas were sorted as in Table 1, the species S. haselowania came next to S. tigrina in descending order. The adaxial epidermal cell surface areas of S. tigrina specimens ranged from 4208.78μ2 to 4259.11μ2 (data not shown). These data did not differ (p=0.05) with those in C29 S. haselowania for the adaxial epidermal cell surface areas (mean: 4215.97 μ2) (Fig. 1). However, the abaxial epidermal cell surface areas of S. tigrina specimens ranged from 2816.88 μ2 to 2917.18 μ2. At the same time, mean abaxial cell surface area of S. haselowania was greater (mean=3556.07 μ2; p<0.05) than that in S. tigrina.
It means that C29 S. haselowania differentiated from plants of S. tigrina. As in this example, there were many other cases where species differences arose from one side of the leaf only.

Fig. 1. Normalized curves for leaf epidermal cell surface areas of S. tigrina, S. haselowania and S. nigroviolacea. a) Adaxial. b) Abaxial. Notice that S. tigrina cannot be differentiated from S. haselowania by their adaxial cell surface areas. However, the 3 species are well distinguished from their abaxial cell surface areas. Notice the change of scale between panels.
Fig. 1. Curvas normalizadas para superficies de células epidérmicas foliares de S. tigrina, S. haselowania y S. nigroviolacea. a) Adaxial. b) Abaxial. Note que S. tigrina no puede diferenciarse de S. haselowania por su superficie de células adaxiales. Sin embargo, las 3 superficies se distinguen bien por la superficie de células abaxiales. Note el cambio de escala entre paneles.
Likewise, the leaf-printing method was applied to alike species which are frequently confused, such as S. tigrina and S. nigroviolacea. Normal standard curves for the adaxial and abaxial epidermal cell surface areas are shown in Figs. 1a and 1b, respectively, for these two species. Mean epidermal cell surface areas were significantly different (p= 0.05) between these two species (Table 2).
Table 2. Statistical comparisons of C15 S. nigroviolacea with samples of S. tigrina. In the "Mean" column, values are the average of n=200.
Tabla 2. Comparaciones estadísticas de S. nigroviolacea C15 con muestras de S. tigrina. En la columna "Promedio", los valores son el promedio de n=200.

Similarly, S. radiosa has been usually confused with S. saccata. However, both species can be distinguished by comparison of their normal standard curves (Fig. 2).

Fig. 2. Normalized curves for leaf epidermal cell surface areas of S. saccata and S. radiosa. a) Adaxial. b) Abaxial. Note the change of scale between panels.
Fig. 2. Curvas normalizadas para superficies de células epidérmicas foliares en S. saccata y S. radiosa. a) Adaxial. b) Abaxial. Note el cambio de escala entre paneles.
Prediction test of unknown samples. Leaf epidermal cell surface areas of six specimens (C03, 26, 33, 53, 77 and 83), received as unknown samples, were analyzed with the leaf printing method. Results obtained with this method were compared with those of already known sample data to make an identification prediction.
Data from samples C26 and C33 matched data from S. tigrina, clones C07 and C59. These results suggest that the unknown samples would correspond to S. tigrina specimens. This prediction was later confirmed when plants flow-ered. Statistical analysis of epidermal cell surface area data showed that neither C07 nor C59 differed significantly (p=0.05) from C26 and C33 samples. Since plants of S. tigrina were imported from Colombia, Guatemala, and Mexico, these data demonstrate that this species does not vary significantly with latitude.
Data from the adaxial and abaxial leaf epidermis of the unknown sample C53 matched (p>0.05) data from the confirmed members C04 and C49, allowing to predict the unknown specimen as S. hernandezii. Two years later, such prediction was confirmed after flowering of the specimen.
Unknown sample C77, received from Chiapas, Mexico was predicted to be S. oculata based on leaf print data. Afterwards, the identification as such species was confirmed when the specimen flowered. Similarly, data from sample C03 matched data S. insignis, which was subsequently confirmed after flowering.
Specimen C83 was received from an unknown origin, and predicted as a member of the Central American group on the basis of its adaxial and abaxial leaf cell surface areas. When C83 flowered, photographs were sent to orchid taxonomists at the Marie Selby Orchid Identification Center (Sarasota, Florida). They determined that it looked like a very yellow form of S. wardii, despite this species is unknown in Central America. The standard normal curves of the adaxial and abaxial epidermal cell surface areas differed significantly (p=0.05) between S. amoena and S. wardii (Table 3). The unknown specimen C83 could not then be a sample of S. wardii. Instead, it fits well with the species from Costa Rica, S. amoena. As a result, specimen C83 was self pollinated to produce seedlings. This would allow to compare vegetative and floral data of the unknown sample with S. amoena. Research to determine the origin of specimen C83 is ongoing.
Table 3. Statistical comparisons of adaxial and abaxial epidermal cell surface areas of S. amoena with samples of S. wardii. Data at each line of the column "Mean" come from n=200.
Tabla 3. Comparaciones estadísticas de superficies de células epidérmicas adaxiales y abaxiales de S. amoena con muestras de S. wardii. Los datos en cada línea de la columna"Promedio" son de n=200.

Phylogenetic aspects. Abaxial and adaxial epidermal cell surface areas were paired in descending order in Table 1. This table was also divided into 3 groups according to the geographical habitat of each species, as it has been reported coming from South America, Central America or Mexico. The combined adaxial and abaxial epidermal cell surface area data for all samples showed species grouped with alike species. Table 1 also offered a pattern for the entire genus. This pattern was broadly divided into three general groups: species coming from either Central or South America or Mexico.
An evolutionary hierarchy was also observed as species data were displayed from larger to smaller epidermal cell surface areas (Table 1). All primitive species exhibited large abaxial and adaxial epidermal cell surface areas. As mean epidermal cell surface areas decreased in the hierarchy, species were arranged by their flowers in a general order as being either primitive or advanced. The primitive species of the genus (S. reichenbachi-ana, S. embreei, S. platyceras, S. grandiflora, S. pulla, S. pozoi, and S. candida) were all from South America.
The mean leaf epidermal cell surface areas of S. oculata placed this species within the Central American group. However, S. oculata can be found in a few scattered locations from northern South America to Southern Mexico, because it is considered a weedy species. Stanhopea reichenbachiana was found only in Colombia while at the northern extreme, S. intermedia was only found in Mexico. Results presented a clear pattern of the evolutionary expansion of the study genus which correlated with declining both adaxial and abaxial leaf epidermal cell surface areas.
DISCUSSION
Morphological data are of significance in systematics because morphological variation can make each taxon unique and distinguishable from each other. Morphometrics and systematic biology share a common interest in the analysis of morphology, when assessing the nature of morphological variation. Biological morphologists should then regard on morphometric tools as an integral part of their approaches to systematics (MacLeod, 2002). In this sense, Stanhopea species show mean adaxial and abaxial leaf epidermal cell surface areas in narrow ranges, separated by wide-enough gaps as to permit species identification. This makes a substantial contribution to the classical botanical approach of needing a floral description to define a species. While it is an evolutionary sine qua non postulate that the primordial flower evolved from a leaf, it is equally obvious that once this divergence took place, natural selection pressures became different for flower and leaves. It means that defining a species by solely analyzing flowers may in fact offer only a sample of organ evolution while distorting the phylogenetic relationships of the species.
The hypothesis that the identification of an unknown species could be statistically determined by comparing its mean leaf epidermal cell surface areas with those of another known species was accepted. It was demonstrated and confirmed by flower analysis in a sufficient number of species. Despite epidermal cell surface areas are not fixed values, they are relatively constant within a narrow range for each species. This allows a correct identification even though some degree of overlapping with closely related species is possible. The identification predictions made on several plant species received as "unknown" which were later confirmed after flowering, proved the accuracy on the applicability of the proposed method. A good example where this technique was proven to be accurate in species confirmation was in the case of S. tigrina and S. nigroviolacea. These two species have been confused by botanical and horticultural authorities during a long time (Williams, 1894; Wilson, 1921; Arnold, 1928; Williams, 1951; Sander et al., 1996). All mentioned S. nigroviolacea as a variety of S. tigrina, despite Beer's (1854) and Kennedy's (1977) valid publications. Stanhopea nigroviolacea is a purely Mexican species, while data from the adaxial and abaxial epidermal leaf cell surface areas inferred S. tigrina to be, although pandemic, from South American origin.
Another case is S. radiosa vs. S. saccata, both species known as coming from Mexico. These species have been confused by taxonomists during decades (Dodson, 1963; Kennedy, 1975). Descriptions were made based on individual flowers, without regard to the whole plant as the species unit which was subjected to a genus variation over a geographical range.
In this investigation, the statistical confidence limit was set at p = 0.05. However, depending on either the material being measured or the level of measurement accuracy, a worker might prefer to select a more rigor-ous test. This statistically-based system using leaf prints is simple, ecologically friendly, and reproducible. Storage of equipment and collected material require a small box for field sampling; small cases for holding microscope glass slides; a place for storing photographs; space for computer equipment, and a microscope. However, best results may be possible by combining the microscope with digital recording and image analysis linked to the computer spreadsheet. With this technological improvement, a larger cell number might be scanned with greater accuracy, results could be offered immediately, and human errors would be minimized. Usefulness of this technique depends partially on the number of samples available for each individual species and the species number sampled throughout the entire range of the genus. This is in order to obtain more accurate results.
The amount of shrinkage observed in Stanhopea leaves is under investigation by comparing data from fresh leaves with similar data from dry leaves. Primitive, large-cell plants appear to show a greater percentage of shrinkage than the smaller-cell, advanced members. Cell shrinkage appears to progress at a relatively uniform rate from primitive to advanced species. Furthermore, mean epidermal cell surface areas vary evolutionarily with speciation of Stanhopea plants with latitude. This has offered new information about this plant genus. If indeed, this applies to other plant genera and families, new knowledge may be gained about the evolution of plants.
REFERENCES
1. Arnold, R.E. (1928). Some Stanhopea species. Orchid review 36: 138-142. [ Links ]
2. Beer, J.G. (1854). Species elevated to specific rank, as Stanhopea nigroviolacea (Morr.). Pract. Stud. Orch. 313 [ Links ]
3. Curry, K.J., W.L. Stern & L.M. McDowell (1988). Osmophore development in Stanhopea anfracta and S. pulla (Orchidaceae). Lindleyana 3:212-220. [ Links ]
4. Dodson, C.H. & G.P. Frymire (1961). Preliminary studies in the genus Stanhopea (Orchidaceae). Annals of the Missouri Botanical Garden 48: 137-172. [ Links ]
5. Dodson, C.H. (1963). The Mexican Stanhopeas. American Orchid Society Bulletin 32: 115-129. [ Links ]
6. Dodson, C.H. (1975). Clarification of some Nomenclature in the genus Stanhopea (Orchidaceae). Selbyana 1: 46-55. [ Links ]
7. Ferry, R.J. Sr., R. Foroughbakhch, L.A. Hauad, S. Contreras, J.V. Star, M.H. Badii & H. Gamez (1997). Leaf-print analyses: an ecologically friendly methodology for plant identification. SIDA 17: 681-690. [ Links ]
8. Kennedy, G.C. (1975). The Stanhopeas of Mexico. Orchid Digest 39: 178. [ Links ]
9. Kennedy, G.C. (1977). Two Confused Stanhopea Species, Stanhopea tigrina and Stanhopea nigrovi-olacea. Orchid Digest 41: 219. [ Links ]
10. MacLeod, N. (2002). Phylogenetic signals in morphometric data. In: MacLeod, N. and Forey, P. L. (eds.), pp 100-138. Morphology, shape and phylogeny. Taylor & Francis, London. 308 p. [ Links ]
11. Sander, C.F, F.K. Sander & L.L. Sander (1996). Sander's List of Orchid Hybrids. 5 year Addendum, 1991-1995. London: The Royal Horticultural Society. 1070 p. [ Links ]
12. Williams, B.S. (1894). The orchid grower's manual. Victoria and Paradise Nurseries, London. 796 p. [ Links ]
13. Williams, L.O. (1951). The Orchidaceae of Mexico. Tegucigalpa, Honduras: CEIBA 2 (1-4), Escuela Agricola Panamericana. 321 p. [ Links ]
14. Wilson, G. (1921). Stanhopea tigrina. Orchid Review 29: 69-70. [ Links ]














