Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO  uBio
uBio
Compartilhar
Phyton (Buenos Aires)
versão On-line ISSN 1851-5657
Phyton (B. Aires) vol.78 no.2 Vicente López jul./dez. 2009
ARTÍCULOS ORIGINALES
Molecular and cultural analysis of seasonal actinomycetes in soils from Artemisia tridentata habitat
Análisis molecular y cultural de actinomicetos estacionales en suelos del hábitat de Artemisia tridentata
Gonzalez-Franco AC1, L Robles-Hernandez1, A Nuñez-Barrios1, JL Strap2, DL Crawford3
1 Facultad de Ciencias Agrotecnológicas, Universidad Autónoma de Chihuahua, Ciudad Universitaria S/N Campus 1, Chihuahua, Chih., 31310, México.
2 Faculty of Science, University of Ontario Institute of Technology, Oshawa, Ontario, L1H 7K4, Canada
3 Department of Microbiology, Molecular Biology and Biochemistry, University of Idaho, Moscow, ID 83844-3052, USA.
Address Correspondence to: AC Gonzalez-Franco, e-mail: conzalez@uach.mx; fax 52-614-4391845; Phone 052-614-4391844.
Recibido-Received 19.09.2008.
Aceptado-Accepted 14.01.2009.
Abstract. In order to understand the temporal dynamics of actinomycete communities of the rhizosphere of the desert plant Artemisia tridentata (sagebrush), two complementary methods were used. They were: (1) 16S rDNA-based PCR coupled with denaturing gradient gel electrophoresis (DGGE), and (2) an agar plate enumeration methodology in which three different media were used to quantify total bacteria, actinomycetes, and fungi. The objective of this research were: (1) to obtain a comprehensive picture of the structure of actinomycete populations, and (2) their dynamics in the rhizosphere of young and old sagebrush plants during two distinct seasons. PCR-DGGE analysis showed that actinomycete groups were less diverse in rhizosphere soils collected in winter than in bulk soils. On the other hand, rhizosphere soil of A. tridentata young plants (RSYP) collected in spring showed an enrichment of actinomycete diversity and/or selection of unique actinomycete populations. This was not the case with the actinomycete populations detected in the rhizosphere soil of old plants (RSOP) in the same season. In this later case, a shift in the dominant bands on PCR-DGGE gels was observed between seasons. Finally, the enumeration analysis showed that actinomycete counts tended to be higher in spring, while total bacteria counts were higher in winter. Although strong soil type effects were detected, more research in the unexplored environment of sagebrush rhizosphere is necessary.
Key words: Actinomycete populations; Seasonal actinomycetes; PCR-DGGE; Rhizosphere soil; Artemisia tridentata; Desert plants.
Resumen. Para comprender la dinámica estacional de las comunidades de actinomicetos en la rizosfera de la planta desértica Artemisia tridentata (sagebrush), se emplearon dos métodos complementarios: (1) PCR basado en el DNAr 16S acoplado con electroforesis de geles con gradientes desnaturalizantes (DGGE), y (2) la metodología de enumeración en placas de agar en la cual se utilizaron 3 medios diferentes para cuantificar bacterias totales, actinomicetos y hongos. Los objetivos de esta investigación fueron: (1) obtener una mayor comprensión de la estructura de las poblaciones de actinomicetos, y (2) determinar su dinámica en la rizosfera de plantas de sagebrush jóvenes y viejas en dos estaciones del año. El análisis de PCR-DGGE mostró que los grupos de actinomicetos fueron menos diversos en suelos obtenidos en invierno asociados a la rizosfera en comparación a aquellos que no estaban asociados. El efecto de la rizosfera para enriquecer la diversidad de actinomicetos y/o selección de poblaciones únicas de actinomicetos fue observado en suelos de rizosfera de planta joven (RSYP) obtenidos en primavera; dicho efecto fue menor en suelos rizosféricos de plantas viejas (RSOP) de la misma estación. En este último tipo de suelo, se observó un cambio en las bandas dominantes en geles de PCR-DGGE entre estaciones. Finalmente, el análisis de plaqueo mostró que el número de actinomicetos fue mayor en primavera, mientras que el de las bacterias totales fue mayor en invierno. Aún cuando se detectó un fuerte efecto del tipo de suelo, se generó información relevante que urge más investigación en el ambiente inexplorado de la rizosfera de sagebrush.
Palabras clave: Poblaciones microbianas; Actinomicetos estacionales; PCR-DGGE; Suelo rizosférico; Artemisia tridentata; Plantas desérticas.
INTRODUCTION
Actinomycetes, phylogenetically defined as a number of taxa within the high G + C subdivision of the Gram-positive phylum (Embley & Stackebrandt, 1994), are involved in important processes in a wide range of habitats (Goodfellow & Williams, 1983; Gooday, 1990; You et al., 1996). They can degrade a wide diversity of recalcitrant compounds such as lignocelluloses (Crawford, 1978) and many other polymers occurring in soil and litter, and a range of xenobiotic compounds (Goodfellow & Simpson, 1987; Warren, 1996; Schrijver & Mot, 1999; Jarerat & Tokiwa, 2001). Because of their metabolic diversity, actinomycetes are a great source of lytic enzymes, antibiotics and a great deal of other bioactive metabolites (Alderson et al., 1993; Sanglier et al., 1993). Actinomycetes are important rhizosphere inhabitants of many plants, where they enhance plant growth and protect the plant roots against attack by phytopathogens (Doumbou et al., 2002). The rhizosphere soil, defined as the soil adjacent to and influenced by plant root exudates (Rovira, 1965), is a selective habitat for microorganisms. Desert plants often grow under stressful environmental conditions in poor soils, and microorganisms growing in the rhizosphere of such plants have to be competitive and adaptive to this environment. Sagebrush plants are known to produce a wide gamut of phenols and terpenoids with antimicrobial activities (Tan et al., 1998). Thus, the rhizosphere of sagebrush plants is a habitat that deserves close examination of the temporal diversity of its actinomycete populations, the kind of microorganisms that are able to colonize and endure unfavorable growth conditions.
PCR-DGGE methodology has been used with success for differentiating genetic fingerprints of complex microbial communities (Muyzer & Smalla, 1998; Koizumi et al., 2002). DGGE banding profiles represent the major constituents of the analyzed complex microbial community. In order to analyze selected populations, such as the actinomycetes, for diversity in the sagebrush rhizosphere or bulk soil, a nested PCR coupled, with DGGE has been successfully used (Heuer et al., 1997). In this study we used nested PCR-DGGE to assess the diversity of actinomycete populations in the sagebrush rhizosphere versus bulk soils from the same habitat. Temporal dynamics of actinomycetes were also determined in two seasons, winter and spring. A cultivation-based methodology was also used along with the molecular biology technique as a complementary approach for examining cultural populations.
MATERIALS AND METHODS
Soil samples. Six soil samples were collected from a sagebrush community near Lewiston, Idaho (48°28'N 116°59'W, elevation 295m) in late January (Winter samples) and late April (Spring samples) of 2002. Three soil samples from rhizosphere soils [one from a young plant (RSYP) and two from old growth plants (RSOP)] were collected. The young plant was 0.3 m tall and 6.4 mm diameter at the base of the trunk; the old plant one was 2.1 m tall and 152.4 mm diameter at the base of the trunk, and the old plant two was 1.5 m tall and 100 mm diameter at the base of the trunk. The plant ages ranged from about 2 to 20 years. Three non-rhizosphere bulk soils nearby to the sagebrush were also collected. Soil samples were collected in plastic bags and immediately placed on ice for transport (Table 1). Care was taken to avoid cross-contamination of the samples. Rhizosphere soils were collected from soil adherent to plant roots to a depth between 8 and 15 cm, while their counterpart bulk soils were sampled approximately 2 m away from the plants underneath the root zone of any grasses growing on the surface. The textural class and pH of the soils were determined as previously reported (Basil et al., 2004).
Table 1. Soils tested in this study and some properties.
Tabla 1. Suelos analizados en este estudio y algunas de sus propiedades.

Soil microbial counts. Total bacteria, actinomycete and fungal counts associated with each soil sample were determined on the following media: Trypticase Soy Agar (TSA) supplemented with 100 μg/ml of cycloheximide for enumeration of total bacteria, Potato Dextrose Agar (PDA) supplemented with 100 μg/ml of cycloheximide for enumeration of actinomycete populations, and PDA supplemented with 100 μg/ml of carbenicillin for enumeration of fungi. The previous media were determined experimentally based on Goodfellow & Williams (1983) information.
Pure culture and total community soil DNA extraction. Chromosomal DNA was extracted from pure cultures of known actinomycete strains (Table 2) using the UltraCleanTM Microbial DNA Isolation Kit (Mo Bio Laboratories, Inc. Solana Beach, CA) according to the manufacturer's instructions. The culture media used to grow the strains are shown in Table 3. Total community DNA was extracted from all soils using the UltraClean Soil DNA kit (Mo Bio Laboratories Inc. Solana Beach, CA), with a modified procedure from that of the manufacturer's. Since one of the general difficulties with extractions from soil is the efficiency of cell lysis, the modified procedure included a brief heat treatment (1.5 min at 65°C) combined with a subsequent bead beating treatment (Vortexed at maximum speed for 15 min). Extraction tubes were then cooled on ice for 1 min. The remaining steps of the procedure were carried on as manufacturer's instructions. In addition, DNA extractions from 1g of soil were purified using the UltraCleanTM PCR Clean-upTM kit (Mo Bio Laboratories, Inc. Solana Beach, CA) according to the manufacturer's instructions.
Table 2. Actinomycete strains used in DGGE analysis.
Tabla 2. Cepas de actinomicetos empleados para el análisis de DGGE.

Table 3. Culture media used to grow specific actinomycete strains.
Tabla 3. Medios de cultivo empleados para crecer cepas específicas de actinomicetos.

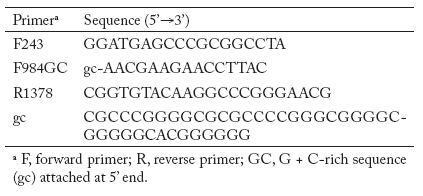
PCR amplifications for actinomycete patterns. Nested PCR was used for amplification of partial 16S rDNA sequences from total soil DNA and genomic DNA isolated from pure cultures of actinomycetes. Primers were chosen to enrich for actinomycetes. The first primer pair was F243- R1378 followed by nested PCR with F984GC -R1378 (Table 4) (Heuer et al., 1997). The first PCR reaction mixture contained 50 mM KCl, 20 mM Tris-HCl, 0.2 mM of each dNTPs, 2.5 mM of MgCl2, 5% (vol/vol) dimethyl sulfoxide, 10 µM of each primer and 2 µl of environmental DNA extract in a 50 µl reaction volume. Dimethyl sulfoxide was added to the reaction mixture to facilitate the denaturation of double-stranded DNA and to circumvent the formation of secondary structures. For amplification of the total environmental soil DNA, 0.1 µg of bovine serum albumin per ml was used to prevent inhibition of the DNA Taq polymerase due to the presence of humic acids (Romanowski et al., 1993). After 5 min of denaturation at 94°C, 1.25 U of Taq DNA polymerase (Invitrogen TECHLINESM, USA) was added at a temperature of 80°C. This hot start was necessary to reduce nonspecific annealing of the primers to nontarget DNA (Chou et al., 1992). The thermocycling conditions were as follows: 94°C for 1 min for one cycle followed by 35 cycles of 94°C for 1 min, 63°C for 1 min, and 72°C for 2 min, and finally one cycle at 72°C for 10 min in a Gene Amp PCR System 2400 thermocycler (Applied Biosystems). Positive and negative controls were included for each PCR experiment. The positive control consisted of reaction mixtures containing 2 µl of genomic DNA of actinomycete pure cultures. The negative control lacked template DNA but contained all other reactants. Four microliters of the PCR products were analyzed on a 1.5% TAE agarose gel stained with ethidium bromide. A 1 Kb plus ladder (Gibco BRL Life Technologies, Gaithersburg, Md.) was used as a DNA size marker. PCR products were excised from a preparative agarose gel and extracted using spin columns (UltraClean GelSpin DNA purification Kit; Mo Bio Laboratories Inc. Solana Beach, CA). The purified DNA was then used as template for a second PCR with primer pair F984GC-R1378 to obtain the fragment 968-1401 suitable for gradient gel electrophoresis analysis. The final PCR products were quantified on a 1.5% agarose gel using a low DNA mass Ladder (Invitrogen, TECHLINESM, USA) and the Gel Doc 1000 fluorescent gel documentation system and Molecular Analyst software (Bio- Rad Laboratories).
Table 4. Primers used in PCR experiments.
Tabla 4. Oligonucleótidos empleados en los experimentos de PCR.
DGGE analysis. The PCR amplicons generated from the soil DNA and genomic DNA of actinomycete strains were separated via DGGE using the Bio-Rad D Gene System according to the manufacturer's instructions. Polyacrylamide (8.3%) gels with gradients between 40% and 75% denaturant (urea-formamide) were prepared; 100% denaturant was defined as 7 M urea and 40% (vol/vol) formamide. The DGGE was performed at 60°C with a standardized voltage of 65 V for 20 h. Approximately 130 ng of each amplified soil DNA was used. After electrophoresis, gels were silver stained (Riesner et al., 1989) to visualize the bands of DNA.
Statistical analysis. Enumerations of the soil microbial populations were statistically analyzed by a pooled ANOVA analysis over the seasons for the six soil samples. This analysis was used to determine if there was an interaction between the independent variables (represented as season x soil) that affects the dependent variable. Interaction terms found to be significant indicate that both independent factors (season and soil) have a combined effect on the dependent variables (e.g. microbial population counts). Significant interaction precludes interpretation of the main effects. All computations were carried out using SAS 8.2 Copyright (c) 1999-2001 by SAS Institute Inc., Cary, NC, USA.
RESULTS
Actinomycete population DNA (PCR-DGGE) analyses. PCR amplifiable DNA was recovered from all the rhizosphere samples, as well as from the corresponding bulk soils. Repeated DGGE analysis of the same PCR product, as well as repeated PCR amplification of the same extracted DNA sample followed by DGGE, produced similar banding profiles, suggesting that the approach was reproducible. In addition, the variation between profiles obtained from replicates was small. Also, separation of 16S rDNA fragments of actinomycete strains spanning the region of positions 968 to 1401 (E. coli positions) were electrophoresed and used as markers (Fig. 1 and 2).

Fig. 1. Separation of 16S rDNA fragments of actinomycete strains spanning the region of positions 968 to 1401 joined to a GC clamp in DGGE; the denaturing gradient was 50-65%. Abbreviations: ssDNA, single stranded DNA; np, Nocardiopsis prasina; sp, Streptosporangium album; ma, Actinomadura cremea subspecies rifanycini; lz, Lentzea californiensis; no, Nocardia peudosporangifera; rh, Rhodococcus facians; ar, Arthrobacter aurescens; ap, Actinoplanes italicus; mm, Micromonospora brunnescens; and st, Streptomyces lydicus.
Fig. 1. Separación de fragmentos del DNAr 16S de cepas de actinomicetos de la posición 968 a la 1401 unidos a una secuencia-abrazadera de GC en DGGE; el gradiente de desnaturalización fue 50-65%. Abreviaturas: ssDNA, DNA de una cadena; np, Nocardiopsis prasina; sp, Streptosporangium album; ma, Actinomadura cremea subespecie rifanycini; lz, Lentzea californiensis; no, Nocardia peudosporangifera; rh, Rhodococcus facians; ar, Arthrobacter aurescens; ap, Actinoplanes italicus; mm, Micromonospora brunnescens; y st, Streptomyces lydicus.
Fig. 2. Analysis of actinomycete community structure for rhizosphere and bulk soil samples collected in spring. DGGE separation of 16S rDNA fragments with actinomycete-specific primers. Sample lanes: (1) RSYP, (2) B1Y, (3) RSOP-1, (4) B1O, (5) RSOP-2 and (6) B2O. Positions of fragments 968-1401 from some actinomycete species are indicated. Abbreviations: np, Nocardiopsis prasina; sp, Streptosporangium album; ma, Actinomadura cremea subspecies rifanycini; lz, Lentzea californiensis; ar, Arthrobacter aurescens; ap, Actinoplanes italicus; and mm, Micromonospora brunnescens.
Fig. 2. Análisis de la estructura de la comunidad de actinomicetos en suelos rizosféricos y no rizosféricos colectados en primavera. Separación de fragmentos de DNAr 16S con oligonucleótidos específicos de actinomicetos a través de DGGE. Carriles de muestras: (1) RSYP, (2) B1Y, (3) RSOP-1, (4) B1O, (5) RSOP-2 y (6) B2O. Se indican las posiciones de los fragmentos 968-1401 de algunas especies de actinomicetos. Abreviaturas: np, Nocardiopsis prasina; sp, Streptosporangium album; ma, Actinomadura cremea subespecie rifanycini; lz, Lentzea californiensis; ar, Arthrobacter aurescens; ap, Actinoplanes italicus; y mm, Micromonospora brunnescens.
To assess whether the rhizosphere, per se, induced shifts in the dominant actinomycete groups in soil, comparisons were made between patterns generated with community DNA from rhizosphere versus bulk soil samples for each season and between seasons.
Analysis of soils collected in winter. The rhizosphere soil profiles from the old and young rhizosphere samples showed less diversity of actinomycetes, as demonstrated by fewer bands in the gel pattern, compared with their counterpart bulk soils (Fig. 2, 3, and 4). The rhizosphere sample of the young plant (RSYP) showed three dominate bands, from which one is only present in this sample compared with its counterpart bulk soil profile, and three weak bands that also were present in the profile of the respective bulk soil. The banding pattern of the bulk soil B1Y showed the highest complexity with five dominating bands and another thirteen weak bands (Fig. 3). Profiles of rhizosphere soil of old plants were similar to those of their respective bulk soils, but with fewer bands and lower intensities, while more dominant bands were detected in the bulk soils (Fig. 3 and 4).
Fig. 3. Comparison of rhizosphere and bulk soil actinomycete profiles of two seasons by PCR-DGGE analysis of 16S rDNA fragment 968-1401. Gel contained 45-75% denaturing gradient. RSOP-1 and B1O sampled in winter (lanes 1 and 2), same soils sampled in spring (lanes 3 and 4), RSYP and B1Y sampled in winter (lanes 5 and 6), same soils sampled in spring (lanes 7 and 8). Dominant bands are indicated by arrows.
Fig. 3. Comparación de los perfiles de actinomicetos de suelos rizosféricos y no rizosféricos presentes en dos estaciones por análisis de PCRDGGE del fragmento 968-1401del DNAr 16S. El gel tiene un gradiente de desnaturalización del 45-75%. RSOP-1 y B1O muestreadas en invierno (carriles 1 y 2), mismos suelos muestreados en primavera (carriles 3 y 4). RSYP and B1Y muestreados en invierno (carriles 5 y 6), mismos suelos muestreados en primavera (carriles 7 y 8). Bandas dominantes se indican con flechas.
Fig. 4. Comparison of rhizosphere and bulk soil actinomycete profiles of two seasons by PCR-DGGE analysis of 16S rDNA fragment 968-1401. Gel contained 50-65% denaturing gradient. B2O and R2O sampled in winter (lanes 1 and 2), same soils sampled in spring (lanes 3 and 4).
Fig. 4. Comparación de los perfiles de actinomicetos de suelos rizosféricos y no rizosféricos presentes en dos estaciones por análisis de PCRDGGE del fragmento 968-1401 del DNAr 16S. El gel tiene un gradiente de desnaturalización del 50-65%. B2O y R2O muestreados en invierno (carriles 1 y 2), mismos tipos de suelo muestreados en primavera (carriles 3 y 4).
Analysis of soils collected in spring. While the profile of the RSYP was more complex than that of its nearby bulk soil, the profiles of the rhizosphere soil of old plants were similar to the profiles of their counterpart bulk soil samples. The pattern of RSYP included six dominant bands, from which one was exclusively found in this sample, and ten weak bands, which also were exclusive to this sample. The pattern of its counterpart bulk soil B1Y, showed less complexity with three dominant bands and six weak bands (Fig. 3). RSOP-1 showed a lower complexity of the banding pattern. It showed 3 dominant bands with four weak bands while that of its counterpart bulk soil B1O was more complex with five dominant bands and nine weak bands (Fig. 3). In the case of RSOP-2 and its bulk soil B2O, showed a similar profile with similar band intensities, having four dominant bands and eight weak ones (Fig. 4).
In both seasons, winter and spring, a large number of bands were shared among the different bulk soil and rhizosphere profiles. Only a few differences were observed, mainly with the RSYP. Even thought the RSYP profile of the sample collected in winter was less complex than the bulk soil, it had one dominant band exclusive to this sample. More differences were observed with the RSYP profile of the samples collected in spring, where a more complex profile and more unique bands were observed (approximately thirteen bands). With rhizosphere soil of old plants, differences were mainly observed as a stronger intensity of bands, but with very low complexity in the profiles compared with their respective bulk soils (Fig. 3). The overall diversity of actinomycete populations was higher in spring than in winter. In the latter, the very low diversity may be attributed to the root exudates of the sagebrush, which may be less diverse, lower in amount and/or more toxic during this time. Earlier studies revealed that sagebrush produces many classes of secondary metabolites, including terpenoids and flavonoids with antibacterial activity; two examples are the artemisinic acid, a precursor for the semi-synthesis of artemisinin, and α-santonin (Tan et al., 1998).
Cultivation based analysis. Cultural microbial analyses were performed to assess and compare microbial counts (total bacteria, actinomycetes, and fungi) within and between seasons. Within seasons, all the microbial counts were significantly higher in all the rhizosphere soils compared to their counterpart bulk soils, except for actinomycete numbers of RSOP-1 collected in Winter which were statistically equal to their counterpart bulk soil B1O (Table 5). A pooled analysis of variance (ANOVA) indicated that there was a significant combined effect of season and soil on the microbial population counts. A per-soil analysis showed that the actinomycete populations were generally higher during spring, except for RSOP-2, which showed actinomycete counts slightly higher in winter (Table 6). On the other hand, the larger total bacterial counts were observed in all the rhizosphere soils in samples collected in winter. These values were statistically different to those from soil samples collected in spring (Table 6). The total bacterial numbers in bulk soils were higher in winter samples for B1Y, no difference between seasons in B2O and higher in spring samples for B2O. Finally, the numbers of fungi were very similar between both seasons (spring and winter), with the exception of counts observed for RSYP and B2O (Table 6).
Table 5. Rhizosphere effect with means of total bacteria, actinomycetes, and fungi per soil within a season.
Tabla 5. Efecto rizosférico en los promedios de bacterias totales, de actinomicetos y de hongos por suelo en cada estación.

Table 6. Means of total bacterial, culturable actinomycetes and fungal counts in rhizospheres and their counterpart bulk soils from an Artemisia tridentata habitat A.
Tabla 6. Promedios de conteos de bacterias totales, actinomicetos cultivables y de hongos en suelos rhizosféricos y sus respectivos suelos no asociados a la raíz de Artemisia tridentata A.

DISCUSSION
The results obtained with PCR-DGGE performed on rhizosphere DNA showed clear profiles that represented the dominant actinomycete fractions in the samples. Their DNA fingerprints showed that there are several dominant groups which are relatively stable in bulk soil and in rhizosphere both in spring and winter season and to a lesser extent between rhizospheres of young and old plants. The potential impact of the root system on actinomycete populations in soil was observed as major shifts within the RSYP. In the latter case, it seems that the main effect of the rhizosphere was exerted on the actinomycete populations, represented by the weaker bands. The overall actinomycete group within the rhizospheres differed between the seasons examined, where in samples collected in spring, showed more complex profiles and/or more intensive dominant bands. The difference in band intensity was presumed to indicate numerical differences between the target molecules. However, although RSOP-2 had a higher diversity and abundance of actinomycete populations in samples collected in spring than in winter, shift in populations were more probably due to soil rather than rhizosphere effects, because the bulk soil showed the same shift between seasons. Gelsomino et al. (1999) observed differences in the grouping of DGGE fingerprints obtained from 16 different soils from different geographical locations, and they found that similar soil types tend to select similar communities. Furthermore, Garbeva et al. (2004) described how small size particle, particularly clay, had a stronger effect over the microbial structure than sandy soils. These findings support the results obtained with RSOP-2, which had a small particle size with higher clay content. Finally, profiles of soil samples collected in winter, showed a clear reduction in actinomycete diversity, probably due to a lower nutrient and/or a more toxic environment at this time of year.
For the culturable microbial analysis within seasons, total bacterial, actinomycete and fungal counts were statistically higher in the rhizospheres compared to their counterpart bulk soil samples, indicating shifts of relative abundance in the soil due to the rhizosphere effect. The rhizosphere effect has been studied and detected in multiple studies, where the input of organic material derived from the plant roots and root exudates plays a major role (Rovira, 1965; Smalla et al., 2001). In the overall analysis of culturable microorganisms, actinomycete counts were higher in samples collected in spring, while total bacterial counts were higher in samples collected in winter. This can be explained by the physiological cycle of the plant. During the spring time, roots are very active, taking up nutrients from the soil. Maximum root growth is known to occur during April (Smalla et al., 2001). Thus, during this period of time, oligotrophic organisms, that is, those that have the ability to multiply and maintain activity in habitats of low carbon flux, such as actinomycetes, will dominate. On the other hand, we hypothesized that in winter, the cycle switches to a resting state and that some of the organic compounds present in the roots are released into the soil, resulting a concomitant increase in the abundance of copiotrophic organisms, which are able to grow in a carbon-rich environment. The same seasonal tendency of actinomycete population abundance of RSYP were detected in both the plate enumeration and the DGGE fingerprints. However, divergences were also detected. For example, RSOP-2 samples, showed the highest diversity of actinomycetes in samples collected in spring, while the culturable number of actinomycetes was greater in winter. Furthermore, actinomycete counts almost all the time were higher in rhizosphere soils rather than in bulk soils independently of the season, while in the DGGE fingerprints such actinomycete abundance was not reflected in many cases. These divergences reflected the picture of relative stability of the structure of the total (culturable plus nonculturable) actinomycete populations contrasted with the picture of variability obtained in the cultivation-based analyses. Then, even though actinomycete populations were more abundant in spring in some cases, no statistically conclusive evidence was obtained, due to the small number of plants analyzed and the soil type effect that introduced variability into the experiments. Therefore, additional experiments with a larger number of plants (considering also soil types) will be required to determine if spring is the best time to collect samples for actinomycete isolates from the rhizosphere of sagebrush. It would be interesting to learn more about population changes during all seasons, in order to gain more insight into the complete picture of the effects of sagebrush roots on the microbial dynamics of this rhizosphere habitat.
ACKNOWLEDGEMENTS
The authors sincerely thank Dr. William Price, Statistician, College of Agriculture, University of Idaho, for his help with the statistical analyses. This research was supported in part by the Idaho Agricultural Experiment Station, and by the "Programa de Mejoramiento del Profesorado (PROMEP)" throughout the "Subsecretaria de Educación Superior e Investigación Científica (SESIC)" of Mexico.
REFERENCES
1. Alderson, G., D.A. Ritchie, C. Cappellano, R.H. Cool, N.M. Ivanova, A.S. Huddleston, C.S. Flaxman, V. Kristufek & A. Lounes (1993). Physiology and genetics of antibiotic production and resistance. Research in Microbiology 144: 665-672. [ Links ]
2. Atlas, R.M. (1997). Handbook of Microbiological Media. Boca Raton, Florida: CRC Press, Inc. [ Links ]
3. Basil, A.J., J.L. Strap, H.M. Knotek-Sinith & D.L. Crawford (2004). Studies on the microbial populations of the rhizosphere of big sagebrush (Artemisia tridentata). Journal of Industrial Microbiology and Biotechnology 31: 278-288. [ Links ]
4. Chou, Q., M. Russel, D. Birch, J. Raymond & W. Bloch (1992). Prevention of pre-PCR mis-priming and primer dimerization improves lowcopy- number amplifications. Nucleic Acids Research 20: 1717-1723. [ Links ]
5. Crawford, D.L. (1978). Lignocellulose decomposition by selected Streptomyces strains. Applied and Environmental Microbiology 35: 1041-1045. [ Links ]
6. Doumbou, C.L., M.K. Hamby-Salove, D.L. Crawford & C. Beaulieu (2002). Actinomycetes, promising tools to control plant diseases and to promote plant growth. Phytoprotection 82: 85-102. [ Links ]
7. Embley, T.M. & E. Stackebrandt (1994). The Molecular Phylogen and Systematics of the Actinomycetes. Annual Review of Microbiology 48: 257-289. [ Links ]
8. Garbeva, P., J.A. van Veen & J.D. van Elsas (2004). Microbial diversity in soil: selection microbial populations by plant and soil type and implications for disease suppressiveness. Annual Review of Phytopathology 42: 243-270. [ Links ]
9. Gelsomino, A., A. Keijzer-Wolters, G. Cacco, & J.D. van Elsas (1999). Assessment of bacterial community structure in soil by polymerase chain reaction and denaturing gradient gel electrophoresis. Journal of Microbiological Methods 38: 1-15. [ Links ]
10. Gooday, G.W. (1990). The ecology of chitin decomposition. Advances in Microbial Ecology 11: 387-430. [ Links ]
11. Goodfellow, M. & S.T. Williams (1983). Ecology of actinomycetes. Annual Review of Microbiology 37: 189-216. [ Links ]
12. Goodfellow, M. & K.E. Simpson (1987). Ecology of streptomycetes. Frontiers in Applied Microbiology 2: 97-125. [ Links ]
13. Heuer, H., M. Krsek, P. Baker, K. Smalla & E.M.H. Wellington (1997). Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Applied and Environmental Microbiology 63: 3233-3241. [ Links ]
14. Jarerat, A. & Y. Tokiwa (2001). Degradation of poly (tetramethylene succinate) by thermophilic actinomycetes. Biotechnology Letters 23: 647-651. [ Links ]
15. Koizumi, Y., J.J. Kelly, T. Nakagawa, H. Urakawa, S. El-Fantroussi, S. Al-Muzaini, M. Fukui, Y. Urushigawa & D.A. Stahl (2002). Parallel characterization of anaerobic toluene- and ethylbenzene-degrading microbial consortia by PCR-denaturing gradient gel electrophoresis, RNA-DNA membrane hybridization, and DNA microarray technology. Applied and Environmental Microbiology 68: 3215-3225. [ Links ]
16. Muyzer, G. & K. Smalla (1998). Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Van Leeuwenhoek 73: 127-141. [ Links ]
17. Riesner, D., G. Steger, R. Zimmat, R.A. Owens, M. Wagenhofer, W. Hillen, S. Volbach & K. Henco (1989). Temperature-gradient gel electrophoresis of nucleic acids: analysis of conformational transitions, sequence variations, and protein-nucleic acid interactions. Electrophoresis 10: 377-389. [ Links ]
18. Romanowski, G., M.G. Lorenz & W. Wackernagel (1993). Use of polymerase chain reaction and electroporation of Escherichia coli to monitor the persistence of extracellular plasmid DNA introduced into natural soils. Microbiol. 59: 3438-3446. [ Links ]
19. Rovira, A.D. (1965). Interactions between plant roots and soil microorganisms. Annual Review of Microbiology 19: 241-266. [ Links ]
20. Sanglier, J.J., H. Haag, T.A. Juck & T. Fehr (1993). Novel bioactive compounds from actinomycetes: a short review (1988-1992). Research in Microbiology 144: 633-642. [ Links ]
21. Schrijver, A.D. & R.D. Mot (1999). Degradation of pesticides by actinomycetes. Critical Reviews in Microbiology 25: 85-119. [ Links ]
22. Shirling, E.B. & D. Gottlieb (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Bact. 16: 313-340. [ Links ]
23. Smalla, K., G. Wieland, A. Buchner, A. Zock, J. Parzy, S. Kaiser, N. Roskot, H. Heuer & G. Berg (2001). Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Applied and Environmental Microbiology 67: 4742-4751. [ Links ]
24. Tan, R.X., W.F. Zheng & H.Q. Tang (1998). Biologically active substances from the genus Artemisia. Plant Medica 64: 295-302. [ Links ]
25. Warren, R.A.J. (1996). Microbial hydrolysis of polysaccharides. Annual Review of Microbiology 50: 183-212. [ Links ]
26. You, M.P., K. Sivasithamparam & D.I. Kurtboke (1996). Actinomycetes in organic mulch used in avocado plantations and their ability to suppress Phytophthora cinnamomi. Biology and Fertility of Soils 22: 237-242. [ Links ]

















