Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
Phyton (Buenos Aires)
versión On-line ISSN 1851-5657
Phyton (B. Aires) vol.79 no.1 Vicente López ene./jun. 2010
ARTÍCULOS ORIGINALES
Association between microsatellites and resistance to Mal de Río Cuarto in maize by discriminant analysis
Asociación entre microsatélites y resistencia a Mal de Río Cuarto mediante análisis discriminante
Bonamico1 NC, MG Balzarini2, AT Arroyo2, MA Ibañez1, DG Díaz3, JC Salerno3, MA Di Renzo1
1 Facultad de Agronomía y Veterinaria, Universidad Nacional de Río Cuarto, Agencia n°3, 5800 Río Cuarto, Argentina.
2 Facultad de Ciencias Agropecuarias, Universidad Nacional de Córdoba, cc 509, 5000 Córdoba, and CONICET (National Council of Scientific and Technological Research), Argentina.
3 Instituto de Genética "Ewald A. Favret", Instituto Nacional de Tecnología Agropecuaria, cc 25, 1712 Castelar, Argentina.
Address Correspondence to: Natalia C. Bonamico, e-mail: nbonamico@ayv.unrc.edu.ar; Phone: 054-0358-4676145.
Recibido - Received 29.IX.2009.
Aceptado - Accepted 25.II.2010.
Abstract. Resistance to Mal de Río Cuarto (MRC) disease in maize (Zea mays L.) is important in Argentina because the crop area involves a wide region where the disease is endemic. Molecular marker-assisted selection could be used as an additional selection tool to enhance precision of the genotype selection for resistance. It demands the identification of informative markers. Microsatellite (SSR) markers linked to gene(s) associated with resistance to MRC have been reported from previous QTL analyses. These analyses have been made on linkage maps derived from a relatively early mapping population. In advanced populations, where highly distinct genotypes are easily classified, discriminant analysis (DA) represents a complementary strategy to marker identification; this method does not require a priori genetic map. The objectives of this study were (1) to identify SSR markers associated with MRC resistance by using DA, and (2) to assess DA-selected SSR markers consistency across environments. The recombinant inbred lines (RILs) were evaluated for disease severity and traits related to symptoms of MRC disease at five environments located in the endemic area. The DNA profiles were obtained using 60 SSR. For discriminant analysis, the RILs were assigned to one of two groups defined to represent low and high values for each trait. A molecular analysis of variance (AMOVA) from marker data found significant molecular differences between the two extreme groups formed for each trait before DA. There was an array of markers associated with the MRC disease severity and with traits related to symptoms of disease. The lack of consistency in the several DA-selected SSR markers across environments indicated that genotype-environment interaction effects were significant. Selected markers can be used to allocate new individuals to predefined groups as well as to infer putative localization of genes with small individual effects on resistance to MRC.
Keywords: Maize; Microsatellite; Mal de Río Cuarto; Discriminant analysis.
Resumen. La resistencia a la enfermedad Mal de Río Cuarto (MRC) en maíz (Zea mays L.) es importante en Argentina debido a que la zona de cultivo abarca gran parte del área donde la enfermedad es endémica. La selección asistida por marcadores podría ser usada como herramienta para incrementar la precisión de selección para resistencia en genotipos de maíz. Ésta requiere la identificación de marcadores informativos. Algunos estudios previos de mapeo de loci de caracteres cuantitativos (QTL) mediante mapas de ligamiento realizados con generaciones tempranas identificaron marcadores microsatélites (SSR) ligados a genes asociados con resistencia a MRC. En generaciones avanzadas la diferenciación de los genotipos permite su fácil clasificación; el análisis discriminante (DA) es un método que no requiere el desarrollo de un mapa genético y representa una estrategia complementaria para la identificación de marcadores. Los objetivos fueron (1) identificar marcadores asociados con resistencia a MRC mediante DA y (2) analizar la consistencia de los SSR identificados a través de ambientes. Las RILs fueron evaluadas para la severidad y los caracteres relacionados a síntomas de la enfermedad Mal de Río Cuarto en cinco ambientes del área endémica. El DNA fue analizado con 60 marcadores SSR. Las RILs fueron asignadas a uno de los dos grupos definidos, por representar bajos y altos valores del carácter mediante el análisis discriminante. Antes de realizar el DA, un análisis molecular de la varianza (AMOVA) mostró diferencias moleculares significativas entre los grupos extremos definidos para cada carácter. Los resultados permitieron identificar un grupo de marcadores asociados con características de resistencia a MRC. La falta de consistencia a través de ambientes en la selección de varios marcadores indica efectos significativos de interacción genotipo-ambiente. Los marcadores seleccionados pueden ser utilizados para asignar nuevos individuos en grupos predefinidos así como para inferir la posible localización de genes con efecto menor sobre la resistencia a MRC.
Palabras clave: Maíz; Microsatélites; Mal de Río Cuarto; Análisis discriminante.
INTRODUCTION
Natural genetic variation is usually due to effects of multiple detectable quantitative trait loci (QTL) (Liu, 1998). Expression of complex traits is the result of the contribution and interaction of numerous genes, each contributing with a small portion to the overall phenotype (Li et al., 2006). Reliable selection of genes with small individual effects is difficult for breeders because of the high trait variation caused by uncontrolled environmental effects. Disease and pest resistance, which are major components of most breeding programs, are usually associated to genes with minor individual effects.
Complex trait dissection via QTL analysis, demand the production of large segregating populations, construction of dense linkage maps and phenotyping of quantitative traits. All pre-requisites involve substantial amount of time, money and labor (Zhang et al., 2005). However, many genes controlling pest resistance have been located through association to molecular markers (Xu et al., 1999; Redinbaugh et al., 2005; Lübberstedt et al., 2006; Redinbaugh & Pratt, 2009). Molecular marker-assisted selection represents an additional selection tool to enhance the precision of the selection in breeding.
A series of agricultural applications of discriminant analysis (DA) has suggested another way to combine molecular marker data with phenotypic performance of genotypes to identify meaningful markers (Capdevielle, 2001; Fahima et al., 2002; Aluko, 2003; Mcharo et al., 2004, 2005; Zhang et al., 2005; Alwala, 2007). Discriminant analysis was first used to identify RAPD markers associated with disease resistance in rice (Capdevielle et al., 2000). Then, it was extended to other marker types such as microsatellite markers (Zhang et al., 2005), and AFLP markers (Capdevielle, 2001; Capdevielle et al., 2001; Mcharo et al., 2004; Alwala, 2007; Miano, 2008; Miano et al., 2008). The idea of applying DA in this particular context is to identify molecular markers significantly associated with a classification of plant material into two groups of extreme performance regarding an agronomical trait. The DA based approach has been regarded as complementary to genetic studies involving QTL mapping, as indicated by comparison of chromosomal location of markers identified using DA and QTL analysis in several mapping populations. The main difference between DA and QTL analysis is that the latter identifies markers linked to gene(s) of interest whereas DA, being multivariate in nature, identifies an array of markers highly associated with the trait(s) of interest. A function of these markers could be used for genotype classification, i.e. to allocate new individuals to a predefined group.
In Argentina, the production of maize for grain is highly affected by Mal de Río Cuarto (MRC) disease, caused by a virus member of the family Reoviridae, genus Fijivirus and transmitted by the planthopper Delphacodes kuscheli Fennah (Homoptera: Delphacidae) vector (Nome et al., 1981; Ornaghi et al., 1993). Mal de Río Cuarto resistance is a quantitative trait with moderate heritability ranging from 0.44 to 0.56 (Di Renzo et al., 2002). Genetic studies on MRC disease have been made using traditional QTL mapping with early-generation F2:3 (Di Renzo et al., 2004). Recombinant inbred lines (RILs) differ from early-generation populations in that they undergo multiple rounds of meiosis before lines are reached. Genotypes with larger homozygosis allow better genetic differentiation. As a result, markers associated with a trait have a greater probability of suggest distinct genotypes (Burr & Burr, 1991). Therefore, the identification of molecular markers significantly associated with a classification of lines into groups of extreme performance via DA, may be an adequate strategy. The application of DA has not been hitherto applied in studies for resistance to MRC disease in maize.
Our objectives were (1) to identify SSR markers associated with MRC resistance by using DA, and (2) to assess DAselected SSR markers consistency across environments. Identification of molecular markers associated with groups of lines differing in phenotype performance regarding a plant disease could suggest the putative localization of genes with small individual effects on resistance.
MATERIALS AND METHODS
Genetic material. A susceptible dent line, Mo17, and a partially resistant red flint line, BLS14 were used as parents to produce a RILs population. BLS14 was developed by selfing at the Instituto Nacional de Tecnología Agropecuaria, Castelar, from the Argentina local variety "Colorado La Holandesa". This is an open pollinated variety. Early-generation populations from the same cross have been previously used for studies of inheritance and mapping of QTL for resistance to MRC disease (Di Renzo et al., 2002, 2004). A group of 144 RILs was developed by self-pollinating a random sample of F2 plants by single seed descent method to the F2:6 generations.
Field evaluation. For disease evaluations, trials were grown in the area where MRC disease is endemic from the temperate semi-arid region (Río Cuarto, Argentina). Trials were conducted during 2004, 2005 and 2006 at the Río Cuarto (33° 8' S, 64° 20' W, 334 masl) location and during 2004 and 2005 at the Sampacho (33° 19' S, 64° 42' W, 510 masl) location. The year-location combinations were regarded as different environments: Río Cuarto 2004 (R4), Río Cuarto 2005 (R5), Río Cuarto 2006 (R6), Sampacho 2004 (S4) and Sampacho 2005 (S5). The experimental design was a randomized complete block design with two plots/RILs at each environment. Each trial included entries of Mo17 and BLS14 as parental controls.
At the beginning of male flowering, 60-70 days after seeding, the RILs were evaluated for disease severity and nine traits related to common symptoms of MRC disease. Individual plants were at first evaluated for each trait, and data averaged at each environment for each RILs. Plants were rated on a discrete scale whose values increased according to the increase of the disease severity. The following traits and rating scales (in parenthesis) were used: (1) superior internodes (0=normal; 1=shortened); (2) flag leaf length (0=normal; 1=shortened); (3) flag leaf width (0=normal; 1=narrow); (4) leaf border (0=normal; 1=serrated); (5) tassel type (0=normal; 1=reduced flower number but fertile flowers; 2=reduced flower number and sterile flowers; 3=no flowers; 4=no tassel); (6) presence and type of enations (0=no enations; 1=mild enations; 2=enlarged veins; 3=galls); (7) "Hockey pole" ears (0=normal; 1="hockey pole" ear; 2=no ear); (8) multiple ears (0=normal; 1=multiple ears; 2=no ear); (9) ears with few or no kernels (0=normal; 1=two thirds of the ear with kernels; 2=one third of the ear with kernels; 3=ear with no kernels; 4=no ear). As a global metric of MRC resistance we use a disease severity score based on a 0 to 3 grade scale proposed by Ornaghi et al. (1999), where 0=no symptoms; 1=mild symptoms: presence of enations on the abaxial side leaves; 2=shortened superior internodes, enations and "hockey pole" ears; 3=severe dwarfing, enations, and small ears with few or no kernels.
Genetic markers. Total genomic DNA was extracted from young expanded leaves of each RIL using the procedure of Saghai-Maroof et al. (1984). In order to improve the purity of the genomic DNA, we used polyvinylpyrrolidone (PVP40) to eliminate hydroxybenzene during the extraction process. Polymerase chain reaction (PCR) amplification methods were carried out using the procedure described by Di Renzo et al. (2004) and products were resolved in a 4% (w/v) super-fine resolution agarose (BMA-FMC BioWhittaker, Rockland, ME USA) gel. DNA profiles were obtained for each RILs, using 60 SSR markers across eight maize chromosomes (1, 2, 3, 4, 6, 8, 9 and 10). The primer sequences were downloaded from the Maize Genetics and Genomics Database (MaizeGDB, http://www.maizegdb.org) and synthesized by Alpha DNA Company, Montreal, Canada (http://www.alphadna.com).
Data analysis. Pearson correlations (r) between the disease severity and the other traits related to symptoms of MRC disease were calculated by the PROC CORR procedure (SAS Institute ver. 9.1). For discriminant analysis, RILs were assigned to one of two groups defined to represent low and high values for each trait. The two extreme phenotypic groups for each trait represented the 1st and 4th quartile of the trait distribution. Missing marker data, which were around 10-15%, were computed using the multiple imputation procedure of SAS. To test presence of variation at the molecular level between predefined phenotypic groups regarding SSR profiles, the analysis of molecular variance (AMOVA) method (Excoffier, 1992) was performed. Before performing DA, we ran a marker selection procedure with PROC STEPDISC (SAS Institute ver. 9.1) using the forward option as selection method, with the select option set to 0.15. The analytic procedure used here is fully detailed in Zhang et al. (2005). Using the selected markers, a non-parametric method (k-nearest-neighbor) of DA was performed within PROC DISCRIM (SAS Institute ver. 9.1). The linear parametric DA (Fisher, 1936) is also recommended because the high robustness in front of outliers, and non-normal or heteroscedastic data. Percentage of correct classification was calculated from cross-validation error rates by using the crossvalidate option within PROC DISCRIM. A high level of correct classification was used to infer an association between molecular markers and agronomic data for a trait expression.
RESULTS
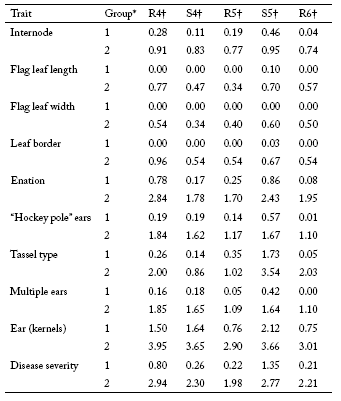
The maize RILs evaluated in this study exhibited a wide range of phenotypic variation for disease severity and all MRC symptoms evaluated. Mean values for the two extreme phenotypic groups for each trait at each environment are shown in Table 1. The phenotypic mean values of the high and low groups were significantly different for all traits in all environments (p<0.001).
Table 1. Mean scores for disease severity and traits related to symptoms of Mal de Río Cuarto in maize for two groups of RILs at five Mal de Río Cuarto endemic environments.
* Group 1: low symptoms; group 2: high symptoms.
† R4, Río Cuarto 2004; S4, Sampacho 2004; R5, Río Cuarto 2005; S5, Sampacho 2005 and R6, Río Cuarto 2006.
Tabla 1. Valores medios de la severidad de enfermedad y los caracteres relacionados a síntomas de Mal de Río Cuarto para dos grupos de RILs de maíz, en cinco ambientes del área endémica.
* Grupo 1: bajo valor medio; grupo 2: alto valor medio.
† R4, Río Cuarto 2004; S4, Sampacho 2004; R5, Río Cuarto 2005; S5, Sampacho 2005 y R6, Río Cuarto 2006.
The molecular variance analysis found significant molecular differences between the two extreme groups for each trait. Table 2 shows the number of markers selected by the STEPDISC procedure applied before DA, and the percent of correct classification of RILs reached with the discriminant function based on the selected markers. For evaluations in R4 and R5 environments, highly percent of correct classification were obtained using all 10 SSR markers. For evaluations in S4 and S5 environments similar results were observed with eleven SSR markers. In the R6 environment, the same percent of correct classification was achieved using among three and nine SSR markers. For disease severity a minimum set of four SSR markers in R4 and S5 environments, and a maximum set of ten SSR markers in R5 environment were selected by DA procedure. Results showed an array of markers associated with the MRC disease severity as well as with traits related to symptoms of MRC disease. The rate of correct classification (obtained by cross-validation) was often higher than 60%.
Table 2. Number of microsatellites pre-selected to classify maize RILs into low and high trait value groups, and percent (%) of correct classification of the discriminant function at five Mal de Río Cuarto endemic environments.
* R4, Río Cuarto 2004; S4, Sampacho 2004; R5, Río Cuarto 2005; S5, Sampacho 2005 and R6, Río Cuarto 2006.
Tabla 2. Número de microsatélites preseleccionados por clasificar las RILs de maíz en los grupos de bajo y alto valor del carácter, y porcentaje (%) de correcta clasificación de la función discriminante en cinco ambientes del área endémica.
* R4, Río Cuarto 2004; S4, Sampacho 2004; R5, Río Cuarto 2005; S5, Sampacho 2005 y R6, Río Cuarto 2006.

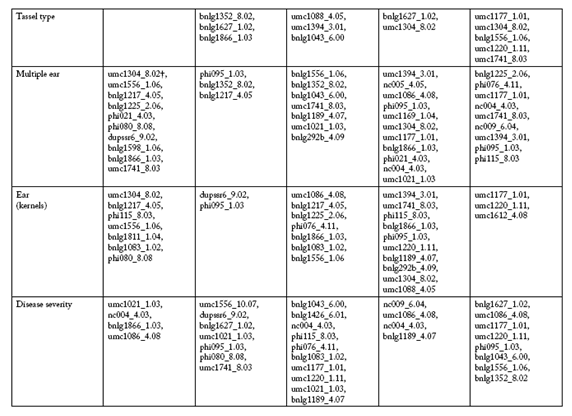
The SSR markers selected by PROC STEPDISC, which differentiate between low and high trait value groups at each environment, are shown in Table 3 and Table 3b. In the R4 environment, the umc1304_8.02, bnlg1217_4.05 and umc1086_4.08 SSR markers were common to five traits, and the bnlg1225_2.06 SSR marker were selected in four of ten traits. In the S4 environment, two SSR markers, bnlg1352_8.02 and phi095_1.03, were identified in five of ten traits. In the R5 environment, the bnlg1189_4.07, bnlg1043_6.00 and phi115_8.03 SSR markers were shared among five traits. In the S5 environment, the bnlg1627_1.02 SSR marker was selected for five traits, while the phi095_1.03 and bnlg1371_6.02 SSR markers were selected for four of ten traits. In the R6 environment, seven and six traits had the umc1177_1.01 and umc1220_1.11 SSR markers in common, respectively.
Table 3. SSR markers pre-selected to classify maize RILs into low and high trait value groups at five Mal de Río Cuarto endemic environments.
* R4, Río Cuarto 2004; S4, Sampacho 2004; R5, Río Cuarto 2005; S5, Sampacho 2005 and R6, Río Cuarto 2006.
† First name-component is SSR marker, second name-component is chromosome and bin number. SSR markers order corresponds to its relative contribution to the discriminant function.
Tabla 3. Marcadores SSR preseleccionados por clasificar las RILs de maíz en grupos de bajo y alto valor del carácter en cinco ambientes del área endémica.
* R4, Río Cuarto 2004; S4, Sampacho 2004; R5, Río Cuarto 2005; S5, Sampacho 2005 y R6, Río Cuarto 2006.
† El primer componente indica el nombre del marcador SSR, el segundo componente indica el cromosoma y el número de bin. El orden de los marcadores SSR corresponde a su contribución relativa en la función discriminante.

In this work, MRC disease severity trait was higher correlated with shortened superior internodes, enations and ears (kernels) traits (r=0.76, 0.79 and 0.70, respectively, p<0.001). The chances of sharing at least one SSR marker increased with stronger correlations. Table 3 shows that in the R4 environment, the bnlg1866_1.03 SSR marker was common to disease severity and shortened superior internodes, while the umc1086_4.08 SSR marker was also shared with enations trait. In the S4 environment, the umc1556_10.07 and umc1021_1.03 SSR markers were selected by DA to disease severity and enations. At the same time, the dupssr6_9.02 and phi095_1.03 SSR markers were identified to disease severity and ears (kernels). In the R5 environment, the bnlg1426_6.01, nc004_4.03 and phi115_8.03 SSR markers were shared between the disease severity and enations traits. The bnlg1083_1.02 SSR marker was common additionally to ears (kernels), and the umc1021_1.03 and bnlg1189_4.07 SSR markers only were detected to disease severity and shortened superior internodes. In the S5 environment, the nc009_6.04 and umc1086_4.08 SSR markers were selected to disease severity and shortened superior internodes, while the bnlg1189_4.07 SSR marker was identified also to ears (kernels). In the R6 environment, the umc1177_1.01 and umc1220_1.11 SSR markers were common to disease severity, shortened superior internodes and ears (kernels). Nevertheless, there were highly correlated traits associated with SSR markers at different positions. For example, in the S4 environment, disease severity and shortened superior internodes characters (r=0.76) showed eight individual SSR markers, but none were shared. Also, there were traits weakly correlated, but sharing common SSR markers. For example, in the R6 environment, disease severity and flag leaf width characters (r=0.28) showed eleven individual SSR markers, with three SSR markers in common.
In relation to assessing DA-selected SSR markers consistency across environments, the umc1021_1.03, nc004_4.03, umc1086_4.08, bnlg 1627_1.02, phi095_1.03, umc1177_1.01, umc1220_1.11, and bnlg1189_4.07 SSR markers were selected by means of DA in two or more environments. For example, the umc1021_1.03 SSR marker was common to the R4, S4 and R5 environments. Likewise, six, ten and eight SSR markers were selected by DA in two of five environments for internodes, enations and ears (kernels), respectively.
DISCUSSION
The genes controlling disease resistance in plants are difficult to identify or characterize precisely. Thus, the need for better forms of disease resistance in agriculture, especially those that hold the promise of long-term durability, calls out to plant pathologists, breeders, geneticists, and molecular biologists to turn their attention to breeding for resistance. Several successful QTLs analysis have been conducted to identify loci controlling disease resistance (Xu et al., 1999; Redinbaugh et al., 2005; Lübberstedt et al., 2006). However, with a few exceptions the DA procedure (Capdevielle et al., 2000) has been applied for this purpose. A series of agricultural applications of DA have suggested a connection between QTL analysis and marker selection to combine molecular marker and agronomic data from cultivar field trials (Capdevielle, 2001; Mcharo et al., 2005; Zhang, 2005; Alwala, 2007; Miano, 2008; Miano et al., 2008). DA-selected SSR markers in this study confirmed such connection. DA allowed predicting phenotypic grouping of maize genotypes of unknown resistance to MRC disease. Even more, a better discriminant function could be built by adding more information about the set of selected markers. The classification of lines with the selected markers for each evaluated traits related to symptoms of MRC disease produced low cross-validation error rates in all environment trials. These results indicate the possibility of using molecular markers to predict phenotypic grouping of new maize genotypes of unknown resistance to MRC disease.
Previous research identified genomic regions with significant effects for MRC resistance located in close proximity to QTL for resistance to either fungal or viral disease. Other maize studies have reported inconsistency in the QTL detected across two independent samples (Ajmone-Marsan et al., 1996; Melchinger et al., 1998). Our results indicated that DA-selected SSR markers pointed to either the same or nearby regions, as previously reported MRC-QTL mapping. Di Renzo et al. (2004), who evaluated an F2:3 population, identified two genomic regions with significant effects for MRC resistance on chromosomes 1 and 8. In a separate study with a different F2:3 mapping population, Kreff et al. (2006) informed on the location of genomic regions with significant effects for MRC resistance on chromosomes 1, 4, 8 and 10. McMullen & Simcox (1995) reported that resistance genes against different pathogens were often clustered in the same chromosomal regions of the maize genome. Redinbaugh et al. (2005) reviewed recent research on virus resistance in maize and suggested that the genes and loci that confer resistance to different viruses tend to be grouped together on just a few chromosomes. Our results suggest that the SSR markers associated with MRC resistance in most of the environments are located on chromosomes 1 and 4.
Disease severity and traits related to symptoms may be caused by common genetic factors. As a large fraction of the observed variation is due to genetic effects, different expressions of MRC resistance with shared SSR markers are expected to correlate; with increasing numbers of shared SSR markers, the correlation may increase. On the other hand, even if shared SSR markers exist, the corresponding traits may be weakly correlated, because these traits may be subject to differential genetic control, or may be differently affected by environmental influences. Recently Lisec et al. (2008) described a similar situation in their study of identification of QTL with Arabidopsis thaliana.
The lack of consistency in several DA-selected SSR markers across environments indicated that genotype-environment interaction effects were significant, as previously reported by Di Renzo et al. (2002, 2004). This observation could also result from the selection of a subset on RILs (genetic drift), loss of alleles during the development of RILs because of an insufficient population size, or due to natural selection. Other work reporting results of QTL analysis from a different genetic background revealed that the identified QTLs were not consistent across environments. Confounding factors such as variability in population structure, sources of parental lines, and different sets of environments could be possible causes for this lack of consistency across environments (Beavis & Smith, 1994). We found that the bnlg1627_1.02, bnlg1371_6.02, bnlg1225_2.06, bnlg1083_1.02, phi080_8.08, and bnlg1556_1.06 SSR markers were often detected across environments. Our results suggest that there are SSR markers associated with disease severity, and traits related to symptoms of MRC disease. These SSR markers could be promising in molecular marker mapping of MRC resistance. This is not a gene mapping study; nevertheless, the markers identified are likely to be associated with QTLs responsible for expression of these traits. Use of discriminant analysis is a complementary platform to QTL analysis (Capdevielle, 2001; Capdevielle et al., 2000; Alwala, 2007; Miano, 2008).
Current research focused on verification of the ability of the selected markers to identify superior maize lines with desirable traits among a group of RILs within a segregating population. Furthermore, several markers were identified via DA; many of them were previously pointed, but others not detected by QTL analysis performed on the early generated linkage map. Results from this work suggest that it is possible to use DA to selected powerful markers that may be useful to breeders. This is a new tool for germplasm improvement providing a discriminant model to integrate the information from markers selected to classify RILs. The model can then be used (1) to facilitate the allocation of new genotypes into groups with distinct performance for MRC resistance, and (2) to identify additional markers associated with the trait. Thus far, results suggested that the complementation of DA and QTL analysis would be a good strategy to identify informative markers.
ACKNOWLEDGEMENTS
This work was supported by grants from Universidad Nacional de Río Cuarto, Agencia Córdoba Ciencia, and Agencia Nacional de Promoción Científica y Tecnológica, Argentina.
REFERENCES
1. Ajmone-Marsan, P., G. Monfredini, A. Brandolini, A.E. Melchinger, G. Garay & M. Motto (1996). Identification of QTL for grain yield in an elite hybrid of maize: Repeatability of map position and effects in independent samples derived from the same population. Maydica 41: 49-57. [ Links ]
2. Aluko, G.K. (2003). Genetic mapping of agronomic traits from the interspecific cross of Oryza sativa (L.) and Oryza glaberrima (Steud). PhD Thesis, Louisiana State University, Baton Rouge, LA, USA. [ Links ]
3. Alwala, S. (2007). Identification of molecular markers associated with resistance to Aspergillus flavus in maize. MSc Thesis, Louisiana State University, Baton Rouge, LA, USA. [ Links ]
4. Beavis, W.D. & O.S. Smith (1994). Identification of quantitative trait loci using a small sample of topcrossed and F4 progeny from maize. Crop Science 34: 882-896. [ Links ]
5. Burr, B. & F.A. Burr (1991). Recombinant inbreeds for molecular mapping in maize: theoretical and practical considerations. Trends in Genetics 7: 55-60. [ Links ]
6. Capdevielle, F.M. (2001). Evaluation of discriminant analysis procedure combining agronomic and molecular marker information for germplasm improvement in rice. MSc Thesis, Louisiana State University, Baton Rouge, LA, USA. [ Links ]
7. Capdevielle, F.M., G.K. Aluko, M. Balzarini & J.H. Oard (2000). Application of molecular markers and discriminant analysis to identify rice lines with contrasting phenotypes for agronomic traits. In: Proceedings of the Fourth International Rice Genetics Symposium, International Rice Research Institute. G. S. Khush, D. S. Brar & B. Hardy (Eds), 216 p. Los Baños, Philippines. [ Links ]
8. Capdevielle, F.M., G.K. Aluko, S.R.M. Pinson, B. Fjellstrom, M. Balzarini, N. Zhang & J.H. Oard (2001). Application of discriminant analysis using molecular markers for classification of rice lines into sheath blight and blast resistance groups: a comparison with QTL interval analysis. In IX Conference, Plant & Animal Genome, 382 p. San Diego, CA, USA. [ Links ]
9. Di Renzo, M.A., N.C. Bonamico, D.G. Díaz, J.C. Salerno, M.A. Ibañez & J.J. Gesumaría (2002). Inheritance of resistance to Mal de Río Cuarto (MRC) disease in Zea mays (L.). Journal of Agricultural Science, Cambridge 13: 47-53. [ Links ]
10. Di Renzo, M.A., N.C. Bonamico, D.G. Díaz, M.A. Ibañez, M.E. Faricelli, M.G. Balzarini & J.C. Salerno (2004). Microsatellite markers linked to QTL for resistance to Mal de Río Cuarto disease in Zea mays L. Journal of Agricultural Science, Cambridge 142: 289-295. [ Links ]
11. Excoffier, L., P.E. Smouse & J.M. Quattro (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479-491. [ Links ]
12. Fahima, T., M.S. Roder, K. Wendehake, V.M. Kirzhner & E. Nevo (2002). Microsatellite polymorphism in natural populations of wild emmer wheat, Triticum dicoccoides, in Israel. Theoretical Applied Genetics 104: 17-29. [ Links ]
13. Fisher, R.A. (1936). The use of multiple measurements in taxonomic problems. Annals of Eugenics 7: 179-188. [ Links ]
14. Kreff, E.D., M.G. Pacheco, D.G. Díaz, C.G. Robredo, D. Puecher, A. Céliz & J.C. Salerno (2006). Resistance to Mal de Río Cuarto Virus in maize: A QTL analysis. Journal of Basic and Applied Genetics 17: 41-50. [ Links ]
15. Li, S.B., Z.H. Zhang, H. Ying, C.Y. Li, J. Xuan, M. Ting, Y.S. Li& Y.G. Zhu (2006). Genetic dissection of developmental behavior of crop growth rate and its relationships with yield and yield related traits in rice. Plant Science 170: 911-917. [ Links ]
16. Lisec, J., R.C. Meyer, M. Steinfath, H. Redesting, M. Becher, H. Witucka-Wall, O. Fiehn, O, Törjék, J. Selbig, T. Altmann & L. Willmitzer (2008). Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. The Plant Journal 53: 960-972. [ Links ]
17. Liu, B.H. (1998). Statistical Genomics: Linkage, Mapping, and QTL Analysis. CRC Press, Inc. Boca Raton, FL, USA. [ Links ]
18. Lübberstedt, T., C. Ingvardsen, A.E. Melchinger, Y. Xing, R. Salomon& M.G. Redinbaugh (2006). Two chromosome segments confer multiple potyvirus resistance in maize. Plant Breeding 125: 352-356. [ Links ]
19. Mcharo, M., D.R. Labonte, J.H. Oard, S.J. Kays & W.L. McLaurin (2004). Linking quantitative traits with AFLP markers in sweetpotato using discriminant analysis. Acta Horticulturae 637: 285-293. [ Links ]
20. Mcharo, M., D.R. Labonte, C. Clark, M. Hoy & J.H. Oard (2005). Molecular marker variability for southern root-knot nematode resistance in sweetpotato. Euphytica 144: 125-132. [ Links ]
21. McMullen, M.D. & K.D. Simcox (1995). Genomic organization of disease and insect resistance genes in maize. Molecular Plant- Microbe Interactions 8: 811-815. [ Links ]
22. Melchinger, A.E., H.F. Utz & C.C. Schön (1998). Quantitative trait locus (QTL) mapping using different testers and independent population sample in maize reveals low power of QTL detection and large bias in estimates of QTL effects. Genetics 149: 383-403. [ Links ]
23. Miano, D.W. (2008). Replication of viruses responsible for sweet potato virus disease in resistant and susceptible sweetpotato genotypes and identification of molecular markers linked to resistance. PhD Thesis, Louisiana State University, Baton Rouge, LA, USA. [ Links ]
24. Miano, D.W., D.R. Labonte & C.A. Clark (2008). Identification of molecular markers associated with sweet potato resistance to sweet potato virus disease in Kenya. Euphytica 160: 15-24. [ Links ]
25. Nome, S.F., S.L. Lenardón, B.C. Raju, I.G. Laguna, S.K. Lowe& D.M. Docampo (1981). Association of reovirus-like particles with "Enfermedad de Río Cuarto" of maize in Argentina. Phytopathologie Zhurnal 101: 7-15. [ Links ]
26. Ornaghi, J., G. Boito, G. Sanchez, G. March & J. Beviacqua (1993). Studies on the populations of Delphacodes kuscheli Fennah in different years and agricultural areas. Journal of Genetics and Breeding 47: 277-282. [ Links ]
27. Ornaghi, J.A., G.J. March, G.T. Boito, A. Marinelli, J.E. Beviacqua, J. Giuggia & S.L. Lenardón (1999). Infectivity in natural populations of Delphacodes kuscheli vector of "Mal Río Cuarto" Virus. Maydica 44: 219-223. [ Links ]
28. Redinbaugh, M.G. & R.C Pratt (2009). Virus Resistance. In: Bennetzen J.L. & S.C. Hake (eds.), pp. 251-270. Handbook of Maize: Its Biology. Springer-Verlag, New York, USA. [ Links ]
29. Redinbaugh, M.G., M.W. Jones & R.E. Gingery (2005). The genetics of virus resistance in maize (Zea mays L.). Maydica 4: 183- 190. [ Links ]
30. Saghai-Maroof, M., K. Soliman, R. Jorgensen & R. Allard (1984). Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proceeding of National Academy of Sciences of the United States of America 81: 8014-8018. [ Links ]
31. SAS Institute Inc. (2004). SAS/STAT User's Guide, Version 9.1. Cary, NC, USA. [ Links ]
32. Xu, M.L., A.E. Melchinger, X.C. Xia & T. Lübberstedt (1999). High-resolution mapping of loci conferring resistance to sugarcane mosaic virus in maize using RFLP, SSR, and AFLP markers. Molecular and General Genetics 261: 574-581. [ Links ]
33. Zhang, N., Y. Xu, M. Akash, S. McCouch & J.H. Oard (2005). Identification of candidate markers associated with agronomic traits in rice using discriminant analysis. Theoretical and Applied Genetics 110: 721-729. [ Links ]














