Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
Phyton (Buenos Aires)
versión On-line ISSN 1851-5657
Phyton (B. Aires) vol.80 no.2 Vicente López jul./dic. 2011
ARTÍCULOS ORIGINALES
Relationships among six herbal species (Curcuma) assessed by four isozymes
Relaciones entre seis especies herbáceas (Curcuma) utilizando cuatro isoenzimas
Deng JB1, CB Ding1, L Zhang1, YH Zhou2, RW Yang1
1 College of Biology and Science, Sichuan Agricultural University, 625014 Yaan, China.
2 Triticeae Research Institute, Sichuan Agricultural University, 611830 Wenjiang, China.
Address Correspondence to: Dr. Yang Ruiwu. College of Biology and Science, Sichuan Agricultural University, 625014 Yaan, China, e-mail: yrwu@sicau.edu.cn
Recibido - Received 22.IV.2011.
Aceptado - Accepted 31.V.2011.
Abstract. Four isozymes, superoxide dismutase (SOD), polyphenol oxidase (PPO), malate dehydrogenase (MDH) and cytochrome oxidase (COD) were studied for identification of six herbal species (Curcuma L.). All the 37 study specimens produced a total of 168 polymorphism isozyme bands. The genetic distance coefficients (GS) varied from 0.08 to 0.54. The dendrogram, obtained according to the polymorphism isozyme bands by the UPGMA method with the software NTSYS-pc2.1, contributed to improve the resolution of phylogeny. From the dendrogram, it was possible to differentiate between the wild and cultivated specimens of C. longa, and within C. sichuanensis species.
Keywords: Curcuma; Isozymes; Genetic relationship; Phylogeny.
Resumen. Se estudiaron cuatro isoenzimas [superóxido dismutasa (SOD), polifenol oxidasa (PPO), málico deshidrogenasa (MDH) y citocromo oxidasa (COD)] para identificar seis especies herbáceas de Curcuma L. Los 37 especimenes estudiados produjeron un total de 168 bandas de isoenzimas polimórficas. Los coeficientes de distancia genética (GS) variaron entre 0.08 y 0.54. El dendrograma, obtenido de acuerdo a las bandas de isoenzimas polimórficas por el método UPGMA con el software NTSYS-pc2.1, contribuyó a resolver la filogenia. A partir del dendrograma, fue posible diferenciar entre los especímenes silvestres y cultivados de C. longa, y dentro de las especies de C. sichuanensis.
Palabras clave: Curcuma; Isoenzimas; Relación Genética; Filogenia.
INTRODUCTION
Curcuma L. (Zingiberaceae) is widespread in the tropics of Asia, Africa and Australia, and it is composed of approximately 70 species (Purseglove, 1974). About 10 Curcuma species are distributed in China (Xiao et al., 1997; Li et al., 2001; Ye et al., 2008). Six of those species have been used as Chinese herbal medicine for more than a thousand years. For example, an extract of their rhizomes exhibits anti-inflammatory, anticancer activity (Moussavi et al., 2006). There are three traditional Chinese medicines [Radix Curcumae (also named Yujin), Rhizoma Curcumae Longae (also named Jianghuang) and Rhizoma Curcumae (also named Ezhu)] derived from these six Curcuma species. Roots of C. longa L.; C. wenyujin Y. H. Chen et C. Ling; C. kwangsiensis S. G. Lee et C. F. Liang, and C. phaeocaulis Valeton (used as herbal species) are officially recorded in Chinese Pharmacopoeia (2010). However, roots of C. sichuanensis C. K. Hsich et H. Zhang, and C. chuanhuanjiang Z. Y. Zhu can also be used as Radix Curcumae; also, the rhizomes of C. chuanhuanjiang, C. sichuanensis and C. wenyujin can be used as Rhizoma Curcumae Longae in folk therapeutic uses (Chen, 1981; Zhu, 1992).
In Traditional Chinese Medicine (TCM) the same medicinal substances can be produced from these six Curcuma species, although one of them can be used as a different medicinal substance. Morphological characteristics are very large for rhizomes and leaves, both intra- and inter-species. The similarities of the growth habit, leaf-shapes, and flowers among the study Curcuma species are so great that it is generally difficult to distinguish the species at both the vegetative and reproductive stages. The Curcuma flowering season vary from April to October, and it is common that the same species has flowers with different colors. These problems have been troublesome in phylogenetic analysis, and made the clinic of TCM inaccurate. However, the correct identity is important to confirm the sources of origin of herbal drugs within the genus Curcuma (Sasaki et al., 2002; Cao & Katsuko, 2003).
Different techniques have different advantages and disadvantages. It is then necessary to confirm the origin of and the genetic relationships among these six herbal species by various methods. Because of the effectiveness of molecular markers in plant systematics (Crawford, 1991), the isozymic technique has been widely applied to (1) deal with evolvement of botany, (2) identify idioplasm resources (Apavatrut et al., 1999; Monireh, 2007), and (3) investigate genetic relationships (Fang et al., 1993; Guo & Li, 2000; Wu et al., 2002; Arzate-Fernandez et al., 2005).
The objectives of this paper were to (1) evaluate the phylogenetic relationships among Curcuma herbal species; (2) explore the taxonomic status of C. sichuanensis and C. chuanhuangjiang species; and (3) identify the origin of traditional medicine in China.
MATERIALS AND METHODS
Plant materials. Thirty seven specimens of the genus Curcuma, which were divided into six species, were analyzed in this study (Table 1). Thirty one specimens were collected from different localities in the Sichuan province, and the other 6 specimens were gathered from the Guangxi Medicinal Botanical Garden. Sichuan and Guangxi belong to well-known regions of these species, and Sichuan is the geo-herbalism habitat of C. longa, C. sichuanensis, C. phaeocaulis and C. chuanhuangjiang in China (Hu, 1998).
Table 1. Origin of the study materials in this research.
Tabla 1. Origen de los materiales estudiados en esta investigación.

Protein extraction and isozyme analysis. For protein extraction, 0.5 g of tender leaves were powdered in liquid nitrogen. Flour of the leaves was first homogenized in the PBS (0.5 mol, pH 7.8) using 1:1 (v/v) ratio, and then centrifuged at - 4 °C for 12 minutes at 12000 rpm. The supernatant was collected and stored at - 20 °C. PAGE was performed according to a modified method of using vertical slab gels (1.5 mm thick) and was set up forming a discontinuous system of two layers. These two layers were: (i) resolving gel: 13.5 cm layer of 7.5% polyacrylamide, and (ii) stacking gel: 1.5 cm layer of 3% polyacrylamide. Four isozymes were tested and the staining protocols followed Wendel & Weeden (1989). Only clear isozyme bands were scored (numbered beginning with the running closer to the origin) and enzymatic schema diagrams painted according to RF values (relative mobility) (Kuhns & Fretaz, 1978). The frequency distribution of isozyme bands was calculated according to enzymatic schema diagram (Tables 2, 3, 4 and 5). Different patterns occurring in each zone of activity (not single bands data) were scored as discrete variables, using '1' to indicate presence, and '0' to indicate absence of a unique pattern. A dendrogram, that depicts the degree of relationships among the taxa, was produced using hierarchical cluster analysis [NTSYS-pc2.1 software (Rohlf, 2000), UPGMA method (Sneath & Sokal, 1973)].
Table 2. Rf value and frequency distribution of PPO bands.
Table 2. Valores Rf y frecuencia de distribución de bandas PPO.

Table 3. Rf value and frequency distribution of COD bands.
Tabla 3. Valores Rf y frecuencia de distribución de bandas COD.
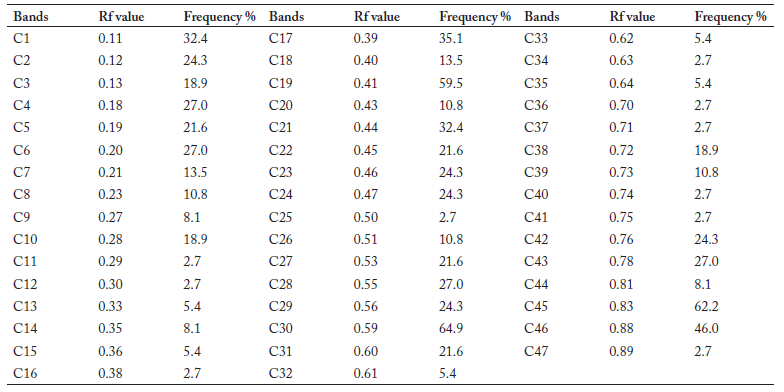
Table 4. Rf value and frequency distribution of SOD bands.
Table 4. Valores de Rf y frecuencia de distribución de bandas SOD.

Table 5. Rf value and frequency distribution of MDH bands.
Table 5. Valores Rf y frecuencia de distribución de bandas MDH.

RESULTS
Isozyme bands. All 37 specimens produced a total of 168 polymorphism isozyme bands; of these, there were 58 PPO, 47 COD, 28 SOD and 35 MDH. Portion pictures of electrophoresis and schema graphs are shown in Figs 1, 2, 3, 4 and 5. Variation of isozyme bands in each specimen were 6-13 (PPO), 6-12 (COD), 2-8 (SOD), and 0-8 (MDH). The isozyme bands showed polymorphism. Consequently, the four isozymes patterns were suitable for fingerprints to distinguish different species of the genus Curcuma.
The bands presented large diversities between species. Two of the C. longa specimens (number 18 and 11) had 6 PPO isozyme bands, and C. phaeocaulis (number 33) produced 13 PPO bands; C. sichuanensis (number 29) had 6 COD bands, and number 12 of C. longa had 12 COD bands; C. phaeocaulis (number 31) had 2 SOD bands, and C. longa (number 1, 2, 5, 7, 8, and 19) had 8 SOD bands; C. wenyujin (number 37) had 8 bands of MDH, and C. phaeocaulis (number 34) had no bands.

Fig. 1. Isozyme band photos of some SOD, PPO, COD and MDH in plants of Curcuma L. A, B: SOD photos of 19-27 and 28-37 materials; C, D: PPO photos of 9-18 and 29-37 materials; E, F: COD photos of 10-18 and 29-37 materials; G, H: MDH photos of 9-18 and 19-27 materials.
Fig. 1. Fotos de las bandas de izoenzimas de algunas SOD, PPO, COD y MDH en plantas de Curcuma L. A, B: Fotos de SOD de los materiales 19-27 y 28-37; C, D: Fotos de PPO de los materiales 9-18 y 29-37; E, F: Fotos de COD de los materiales 10-18 y 29-37; G, H: Fotos de MDH de los materiales 9-18 y 19-27.

Fig. 2. Isozyme patterns of PPO enzyme to identify the 37 specimens of six Curcuma species. (Numbers from 1 to 37 refer to the materials listed in Table 1).
Fig. 2. Modelo de distribución de las isoenzimas de la enzima PPO para identificar los 37 especímenes de las seis especies de Curcuma. (Los números 1 a 37 se refieren a los materiales listados en la Tabla 1).
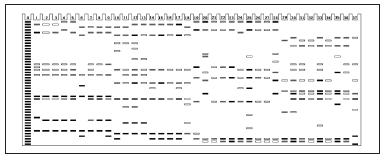
Fig. 3. Isozyme patterns of COD enzyme to identify the 37 specimens of six Curcuma species. (Numbers from 1 to 37 refer to the materials listed in Table 1).
Fig. 3. Modelo de distribución de las isoenzimas de la enzima COD para identificar los 37 especímenes de las seis especies de Curcuma. (Los números 1 a 37 se refieren a los materiales listados en la Tabla 1).
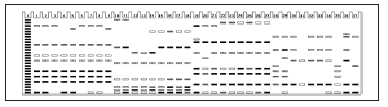
Fig. 4. Isozyme patterns of SOD enzyme to identify the 37 specimens of six Curcuma species. (Numbers from 1 to 37 refer to the materials listed in Table 1).
Fig. 4. Modelo de distribución de las isoenzimas de la enzima SOD para identificar los 37 especímenes de las seis especies de Curcuma. (Los números 1 a 37 se refieren a los materiales listados en la Tabla 1).
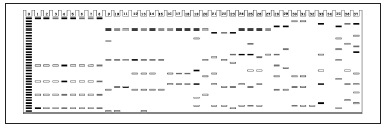
Fig. 5. Isozyme patterns of MDH enzyme to identify the 37 specimens of six Curcuma species. (Numbers from 1 to 37 refer to the materials listed in Table 1).
Fig. 5. Modelo de distribución de las isoenzimas de la enzima MDH para identificar los 37 especímenes de las seis especies de Curcuma. (Los números 1 a 37 se refieren a los materiales listados en la Tabla 1).
Genetic relationships analysis. The Jaccard's similarity coefficients were calculated with the four isozymes data and the genetic distance coefficients (GS) varied from 0.08 to 0.54 among the 37 specimens. The phylogenetic tree (Fig. 6) was constructed following the UPGMA method according to RF values.}
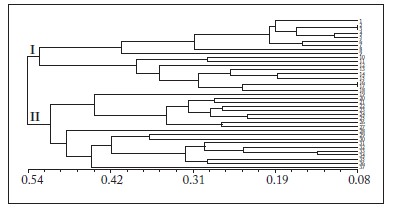
Fig. 6. Dendrogram based on UPGMA analysis of genetic similarity obtained from isozymes data. (Numbers from 1 to 37 refer to the materials listed in Table 1).
Fig. 6. Dendrograma basado en análisis UPGMA de similaridad genética obtenido de los datos de las isoenzimas. (Los números 1 a 37 se refieren a los materiales listados en la Tabla 1).
When GS was 0.52, the 37 specimens were largely divided into two groups. The first group contained 18 specimens of C. longa, all of which were collected from the Sichuan province. The second group included four specimens of C. longa, and all specimens of the other five species. In the first group, most of the cultivated C. longa specimens assembled together as a subgroup and some of the wild C. longa clustered as another subgroup. In the second group, four cultivated C. longa specimens and five C. sichuanensis specimens (numbers 23-27) belonged to the same subgroup; another subgroup included all the specimens of C. phaeocaulis, C. chuanhuangjiang, C. kwangsiensis, C. wenyujin and two cultivated C. sichuanensis species (numbers 28 and 29).
DISCUSSION
When GS was at 0.28, C. kwangsiensis clustered together with one specimen of C. phaeocaulis (number 33); thereafter, they grouped with the other four specimens of C. phaeocaulis. Results indicated that C. kwangsiensis and C. phaeocaulis had relatively closer genetic relationships. However, there might be an error due to the phylogeny tree methods, or this error may be the result of the long-term companion planting in the Medicinal Botanical Garden of Guangxi Autonomous Region. Further DNA analyses are needed to detect this error.
On the study of RAPD marker analysis (Chen et al., 1999), 119 bands were produced with 12 primers, and the genetic distance was 0. 164 between C. wenyujin and C. sichuanensis; the intraspecific genetic distance was much larger than interspecific: they were 0.866 for C. wenyujin and 0.885 for C. sichuanensis. Chen et al. recognized that C. wenyujin was close to C. sichuanensis, and merged C. sichuanensis into C. wenyujin. It is difficult to identify these two species on the level of DNA. While the 18S rRNA and trnK gene sequences of C. sichuanensis and C. longa corresponded completely to the types either 1a or 1b (Sasaki et al., 2002), and the sequence of C. wenyujin belonged to type 5, great differences were shown between C. wenyujin and C. sichuanensis in the trnK and 18S rRNA sequences. C. sichuanensis and C. wenyujin clustered together after clustering study of leaf epidermal features (Xiao et al., 2000). However, Xiao et al. confirmed that C. sichuanensis was close to C. longa rather than C. wenyujin following investigation of the origin and the colour of tuber. It means that former studies of these two species had some different results. Based on our study, the isozymes patterns of PPO, COD, SOD, and MDH showed significant diversities within C. wenyujin and C. sichuanensis; they were included into different groups in the phylogenetic tree. The genetic relationship was remote between C. wenyujin and C. sichuanensis, and C. sichuanensis was close to C. longa.
The relationship between C. longa and C. sichuanensis was complex (Xia et al., 1999; Xiao et al., 1997, 2000, 2001). On the morphological study of leaves and rhizomes, Xiao et al. (2004a, 2004b, 2004c) indicated that C. sichuanensis was the cultivated variety of C. longa. However, they contradicted themselves in their study of leaves and rhizomes: (i) on the morphological study of leaves, C. wenyujin and C. sichuanensis clustered together firstly, and C. longa was far away from them; (ii) on the morphological study of rhizomes, C. longa and C. sichuanensis got together at first. Quan et al. (2005) examined the contents of curdione and turmerol by means of HPLC and 5sRNA sequence on five species (C. kwangsiensis, C. wenyujin, C. phaeocaulis, C. longa, C. sichuanensis). Their results showed that C. longa was on intimate relationship with C. sichuanensis. Tang et al. (2008) recognized that C. sichuanensis was the cultivated mutation species of C. longa by isozyme patterns of POD and EST. In the present study, three cultivated (numbers 23, 24 and 27) and two wild specimens (numbers 25 and 26) of C. sichuanensis were clustered together with four specimens of C. longa at first; two other cultivated specimens (numbers 28 and 29) of C. sichuanensis were gathered with C. chuanhuangjiang, C. phaeocaulis, C. kwangsiensis and C. wenyujin. No doubt that the taxonomic status of C. sichuanensis needs further study.
Although a detailed Flora Sichuanica by Zhu (1992), C. chuanhuangjiang is not mentioned in the Flora of China. Liu & Wu (1999) merged C. chuanhuangjiang into C. kwangsiensis. Xiao et al. (2004a, 2004b, 2004c) thought that C. chuanhuangjiang was the cultivated mutation of C. longa. In our study, the isozymes patterns of C. chuanhuangjiang were dissimilar to the other 36 specimens. Taking into account the previous analysis of Cao & Katsuko (2003) and Tang et al (2008), we believed that it is much more reasonable to retain C. chuanhuangjiang as an individual species.
In conclusion, the four isozymes successfully supported the taxonomical classification of the six Curcuma species. From the dendrogram, 3/4 of the wild specimens of C. longa (numbers 11-18), and the two wild species of C. sichuanensis (numbers 25 and 26), clustered together first in groups I and II, and then gathered with other cultivated specimens; it is shown that the protein differentiation already occurred between cultivated and wild species. We strongly suggest paying attention to the distinction between cultivated and wild specimens when making classification, as well as on the clinic of TCM.
ACKNOWLEDGEMENTS
The project was funded by the Sichuan Youth Science and Technology Foundation (No. 07JQ0085).
REFERENCES
1. Apavatrut, P., S. Anuntalabhochai, P. Sirirugsa & C. Alisi (1999). Molecular markers in the identification of some early flowering Curcuma L. (Zingiberaceae) species. Annals of Botany 84: 529- 534. [ Links ]
2. Arzate-Fernandez, A., C. Meja-González, T. Nakazaki, Y. Okumoto & T. Tanisaka (2005). Isozyme electrophoretic characterization of 29 related cultivars of lily (Lilium spp.). Plant Breeding 124: 71-78. [ Links ]
3. Cao, H. & K. Katsuko (2003). Molecular identification of six medicinal Curcuma plants produced in Sichuan: Evidence from plastid trnK gene sequences. Acta Pharmaceutica Sinica 38: 871-875. [ Links ]
4. Chen, Y.H. (1981). Preliminary study of Curcuma in China. Plant appraisal. Acta Pharmaceutica Sinica 16: 385-385. [ Links ]
5. Chen, Y.H., S.M. Bai, K.D. Chen & S. Zhang (1999). RAPD analysis on Curcuma wenyujin and Curcuma sichuanensis. China Journal of Chinese Materia Medica 24: 131-133. [ Links ]
6. China Pharmacopoeia Committee (2010). Chinese Pharmacopoeia Vol. 1. Beijing Chinese Medicine and Technology Publishing House, pp. 193-194. [ Links ]
7. Crawford, D.J. (1991). Plant molecular systematics: Macromolecular approaches. New York, Wiley and Sons. [ Links ]
8. Fang, D.Q., W.C. Zhang & S.Y. Xiao (1993). Studies on Taxonomy and Evolution of Citrus and its related Genera by Isozyme Analysis. Acta phytotaxonomica Sinica 31: 329-331. [ Links ]
9. Guo, F.G. & Y.H. Li (2000). Identification of Cuscuta L. by Isozyme Analysis. Chinese Traditional and Herbal Drugs 31: 552-553. [ Links ]
10. Hu, S.L. (1998). Chinese genuine traditional Chinese unbleached illustrations. Shandong science and technology publishing house. pp. 250-252. [ Links ]
11. Kuhns, L.J. & T.A. Fretaz (1978). Distinguishing rose cultivars by polyacrylamide gelelectrophoresis II isozyme variation among cultivars. Journal of the American Society for Horticultural Science 103: 509-516. [ Links ]
12. Li, J., D.Z. Zhang & L.X. Gao (2001). The Overview Research of Chinese Radix Curcumae. Nei Mongol Journal of Traditional Chinese Medicine 1: 37-38. [ Links ]
13. Li, X.K., C.S. Yao, Y.D. Huang & J. Xiao (2005). Application of Modern Techniques in Curcuma Reasearch. Pharmaceutical Biotechnology 2: 134-137. [ Links ]
14. Liu, N. & T.L. Wu (1990). The Peroxidase Isozyme of Curcuma L. Guihabia 10: 63-70. [ Links ]
15. Liu, N. & T.L. Wu (1999). Notes on Curcuma in China. Journal of Tropical and Subtropical Botany 7: 146-150. [ Links ]
16. Monireh, C., E. Hassan, S. Azam & N. Vahid (2007). Vahid Niknam Isozyme variation in some populations of wild diploid wheats in Iran. Biochemical Systematics and Ecology 35: 363-371. [ Links ]
17. Purseglove, J.W. (1974). Tropical crops monocotyledons. London: Longman Group Ltd. [ Links ]
18. Quan, X., J.Z. Kui, G.H. Zhao, Z. Ping, T.X.D. Tina, P.L. Shao& W.K.T. Karl (2005). Molecular Genetic and Chemical Assessment of Rhizoma Curcumae in China. Journal of Agricultural and Food Chemistry 53: 6019-6026. [ Links ]
19. Sasaki, Y., H. Fushimi, H. Cao, S.Q. Cai & K. Komatsu (2002). Sequence analysis of Chinese and Japanese Curcuma drugs on the 18S rRNA gene and trnK gene and the application of amplification refractory mutation system analysis for their authentication. Biological & Pharmaceutical Bulletin 25: 1593-1599. [ Links ]
20. Sneath, P. & R. Sokal (1973). Numerical taxonomy: the principles and practice of numerical classification, Vol. 21. San Francisco. [ Links ]
21. Tang, J.Y., Q.M. Li, R.W. Yang, J.Q. Liao & Y.H. Zhou (2008). Study on isozymes in six species of Curcuma. China Journal of Chinese Materia Medica 33: 1381-1386. [ Links ]
22. Tewtrakul, S., S. Subhadhirasakul & S. Kummee (2003). HIV-1 protease inhibitory effects of medicinal plants used as self medication by AIDS patients. Songklanakarin Journal of Science Technology 25: 239-243. [ Links ]
23. Tuchinda, P., V. Reutrakul, P. Claeson, U. Pongprayoon, T. Sematong, T. Santisuk & W. Taylor (2002). Anti-inflammatory cyclohexenyl chalcone derivatives in Boesenbergia pandurata. Phytochemistry 59: 169-173. [ Links ]
24. Wendel, J. & N. Weeden (1989). Visualization and interpretation of plant isozymes: Isozymes in plant biology. Portland, Oregon, Dioscorides Press. pp. 5-45. [ Links ]
25. Wu, W., Y.L. Zheng, L. Chen, R.W. Yang, Z.H. Yan & Y.M. Wei (2002). lsozymes Variations among the Germplasm Resources of Houttuynia in Sichuan. China Journal of Chinese Materia Medica 25: 695-698. [ Links ]
26. Xia, W.J., X.H. Xiao, F.Q. Liu, Z.W. Su & C.Z. Qiao (1999). Determination on Chemical Constituents of Curcuma L. Produced in China. China Journal of Chinese Materia Medica 24: 423-447. [ Links ]
27. Xiao, X.H., F.Q. Liu, C.H. Shi, L.Y. Li, S.Y. Qin, C.Z. Qiao & Z.W. Su (2000). RAPD polymorphism and authentication of medicinal plants from Turmeric (Curcuma L.) in China. Chinese Traditional and Herbal Drugs 31: 209-212. [ Links ]
28. Xiao, X.H., C.Z. Qiao, Z.W. Su, G.P. Yin, Q.M. Fang, G.M. Su, S.Y. Qin, Y. Zhou & L.Y. Li (1998). Recognition technique of the histomorphological images of Radix Curcumae. Chinese Pharmaceutical Journal 2: 14-17. [ Links ]
29. Xiao, X.H., Z.W. Su, C.Z. Qiao & Z.Y. Luo (1997). Advances in the study on medicinal of Curcuma. Chinese Traditional and Herbal Drugs 28: 114-118. [ Links ]
30. Xiao, X.H., W.J. Xia, S.Y. Qin, J.L. Li, Q.M. Fang, G.M. Su & Z.W. Su (2001). Pattern Recognition of Stereoscopic Features of the Leaves Epidermis of Medicinal Curcuma Plants in China by Image Analysis. China Journal of Chinese Materia Medica 26: 523-528. [ Links ]
31. Xiao, X.H., Y.L. Zhao, J. Cheng, G.M. Su, Q.M. Fang & Z.W. Su (2004a). Histological and morphological studies on the rhizomes of Curcuma. China Journal of Chinese Materia Medica 29: 395-399. [ Links ]
32. Xiao, X.H., Y.L. Zhao, J. Cheng, G.M. Su, Q.M. Fang & Z.W. Su (2004b). Histological and morphological studies on leaves of Curcuma in China. China Journal of Chinese Materia Medica 29: 203-207. [ Links ]
33. Xiao, X.H., G.Y. Zhong, G.M. Su, L.Y. Li, Q.M. Fang, S.Y. Chen & Z.W. Su (2004c). Numerical taxonomy of medicinal plants of Curcuma in China. China Journal of Chinese Materia Medica 29: 15-24. [ Links ]
34. Ye, X.B., J. Chen & N. Liu (2008). Curcuma nankunshanensis (Zingiberaceae): A New Species from China. Journal of Tropical and Subtropical Botany 16: 472-476. [ Links ]
35. Zhu, Z.Y. (1992). Flora Sichuanica Vol. 10. Sichuan Nationalities Publishing House. pp. 604-610 [ Links ]














