Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
Phyton (Buenos Aires)
versión On-line ISSN 1851-5657
Phyton (B. Aires) vol.80 no.2 Vicente López jul./dic. 2011
ARTÍCULOS ORIGINALES
Molecular markers to study the variability within the Eragrostis curvula complex
Marcadores moleculares para el estudio de la variabilidad dentro del complejo Eragrostis curvula
Zappacosta D1, M Meier1, A Carrera1, G Pacheco2, S Cardone3, JP Selva1, V Echenique1
1 Lab. Biotecnología Vegetal, CERZOS-CONICET, CCT Bahía Blanca and Departamento de Agronomía, Universidad Nacional del Sur, San Andrés 800, (8000) Bahía Blanca,
Argentina.
2 Instituto de Genética INTA-Castelar.
3 Cátedra de Genética, Facultad de Agronomía, Universidad Nacional de Buenos Aires.
Address Correspondence to: Dr. Viviana Echenique, Departamento de Agronomía, Universidad Nacional del Sur, San Andrés 800, (8000) Bahía Blanca, Argentina. Phone 54 291
4861666, ext. 155; Fax 54 291 4862882, e-mail: echeniq@criba.edu.ar
Recibido - Received 3.VIII.2011.
Aceptado - Accepted 5.VIII.2011.
Abstract. Weeping lovegrass is well adapted for forage production and useful for soil conservation in semiarid regions, constituting a morphologically diverse group. Diploid genotypes are unfrequent and reproduce sexually, whereas the tetraploids and plants of higher ploidy levels reproduce by apomixis. In the present work RAPD, AFLP and EST-SSR were used in order to assess the reproductive mode through progeny tests, to determine intracultivar homogeneity or seed purity, to establish genetic relationships among the cultivars within the complex and to characterize the new materials obtained by our group. Eight commercial cultivars and three new plant materials were analyzed. Uniform and variable patterns were observed in progenies of apomictic and sexual plants, respectively. Seed purity was evaluated in seed bulks, observing a certain degree of contamination with seeds from different sources. AFLP were the markers with the highest potential for cultivar identification. The clustering using SSR and AFLP was consistent with previous studies using isozymes and morphological traits. The new tetraploid materials developed by our own group should be included within the curvula type. We also proposed the creation of a new morphological type for the diploid, because it posses morphological characteristics of robusta and molecular profiles of the conferta type.
Keywords: Eragrostis; Molecular markers; Apomixis; Seed purity; Cultivar identification.
Resumen. El pasto llorón es una especie forrajera que se destaca por su extraordinaria rusticidad, su capacidad para prosperar en suelos pobres en fertilidad y su aptitud para consolidar suelos erosionables. Se trata de un grupo botánico muy polimórfico. Los genotipos diploides son poco frecuentes y de reproducción sexual, mientras que los individuos tetraploides o de mayor ploidía se reproducen por apomixis. En el presente trabajo se utilizaron marcadores moleculares (RAPD, AFLP y EST-SSR) para analizar el modo reproductivo a través de pruebas de progenie, para determinar la homogeneidad intracultivar o pureza de semillas, para establecer relaciones genéticas entre genotipos dentro del complejo y para caracterizar nuevos materiales obtenidos por nuestro grupo. Ocho cultivares comerciales y tres nuevos materiales fueron estudiados. Patrones de amplificación uniformes y variables fueron obtenidos en progenies de plantas sexuales y apomícticas, respectivamente. La pureza de semillas fue evaluada sobre lotes de semillas comerciales, observándose contaminación en algunos lotes. Los AFLP fueron los marcadores con mayor potencial para la identificación de cultivares y los dendrogramas obtenidos usando SSR y AFLP fueron consistentes con estudios previos realizados con isoenzimas y caracteres morfológicos. El nuevo material vegetal tetraploide desarrollado por nuestro grupo de trabajo podría ser incluido dentro del tipo curvula. También se propone la creación de un nuevo tipo morfológico para el diploide, porque posee características morfológicas de tipo robusta y moleculares de tipo conferta.
Palabras clave: Eragrostis; Marcadores moleculares; Apomixis; Pureza de semillas; Identificación varietal.
INTRODUCTION
Eragrostis Wolf, consisting of more than 350 species, is the largest genus among the Eragrostideae, a tribe of Chloridoideae, one of the most diverse grass subfamilies. The tribe is regarded as an unnatural taxonomic group, and Eragrostis itself is a large polyphyletic assemblage (Roodt-Wilding & Spies, 2006). Eragrostis curvula (Schrad.) Nees (weeping lovegrass) is a vigorous grass native to Southern Africa that is well adapted for forage production and useful for soil conservation in semiarid regions of several countries, such as US, Argentina, South Africa and Australia. This species constitutes a morphologically diverse group, poorly understood and not well circumscribed. For these reasons, it is frequently referred as E. curvula complex or E. curvula sensu lato (Voigt et al., 2004). It contains intermediate forms that overlap with E. chloromelas and E. lehmanniana, and for that reason some authors have proposed that these species should be included in E. curvula sensu lato (de Winter, 1955). Although there are clear morphological differences between varieties, the existence of ecotypes with intermediate phenotypes hinders classification (Poverene & Voigt, 1997). Plants originated via intra or inter-specific crosses combine morphological and agronomic traits of the parental varieties.
Six agronomic types were initially described (Leigh, 1961; Leigh & Davidson, 1968), namely curvula, robusta blue, robusta green, robusta intermediate, tall chloromelas and short chloromelas. An additional group called conferta was later recognized (Jacobs, 1982). This classification was principally based on morphological traits of leaves and inflorescences, plant size and growth habit. More recently, Covas (1991) proposed a new morphological classification for materials growing in Argentina based on seeds, leaf blades and panicle characteristics. However, such characters are influenced by growth conditions and fertilization, and although they are generally distinguishable in mature plants, they are difficult to recognize at plantlet stage. The high morphological diversity existing in this complex is accompanied by a similar variability in physiological characteristics and agronomic traits such as cold resistance, photoperiod response, adaptation to various soil types, forage quality and grazing response (Voigt et al., 2004). The morphological types and the cultivars most frequently used in Argentina are the following: curvula (cvs. Tanganyika, Ermelo, Morpa and Don Arturo INTA - Instituto Nacional de Tecnología Agropecuaria), pilosa (cv. Don Juan INTA), conferta (cv. Don Walter INTA) and robusta (cvs. Don Carlos INTA, Don Pablo INTA and Don Eduardo INTA).
Eragrostis curvula comprises a classic agamic complex, as was described by Harlan and de Wet (1963) where diplosporous apomixis followed by pseudogamy is the prevalent reproductive mode (Voigt & Bashaw, 1972). Sexual plants are rare and hybridize with apomictic forms to produce new apomictic ecotypes, which account for most of the variability in the complex. Chromosome number of the species ranges from 2n=2x=20 to 2n=8x=80 (Vorster & Liebenberg, 1977). Within the family Poaceae, intraspecific variation in chromosome number is very common (Quarín et al., 1996). Diploid genotypes are unfrequent and display a sexual reproductive mode, whereas the tetraploids and higher level ploidy plants reproduce by apomixis.
In plant breeding programs, information on the genetic diversity within and among closely related crop species is essential for a rational use of genetic resources. It is particularly useful in characterizing individual accessions and cultivars, in detecting duplications of genetic materials in germplasm collections, and as a general guide in selecting parents for crossing in breeding programs and mapping. For these purposes molecular markers are very useful tools, representing interesting alternatives for solving problems of varietal identification and for providing information to complete the morphological and biochemical characterization.
AFLP, RAPD, ISSR and RFLP have been used for the characterization of cultivars and accessions of different plant species. Within Eragrostideae, these markers were used for molecular characterization and mapping of the major cereal from Ethiopia, Eragrostis tef (Bai et al., 2000; Yu et al., 2007). This is a sexual autotetraploid (2n=2x=40) species with a relatively small genome, compared with other cereals, with a 2C DNA content ranging from 1.48 to 1.52 pg and a haploid genome size between 714 and 733 Mbp, (Ayele et al., 1996), close to the estimated for E. curvula (Bennet & Smith, 1976). In E. curvula, biochemical markers (isozymes) have been used for the characterization of cultivars and the assessment of the reproductive mode (Poverene & Voigt, 1997).
Our group has recently carried out in vitro culture assays using E. curvula materials with different reproductive modes and ploidy levels for breeding and research purposes. A sexual diploid plant (2x sex) derivative from cv. Tanganyika (4x apo) was obtained by inflorescence in vitro culture and then two highly sexual tetraploid plants (4x sex) were originated from the duplication of the diploid one using colchicine (Cardone et al., 2006). Different approaches were then designed in order to characterize the novel materials. RAPD and AFLP were used in progeny tests (Cardone et al., 2006) and to study the genetic structure of the euploid series obtained (Mecchia et al., 2007). EST-SSRs were also developed from cDNA libraries of leaves and panicles of this plant series (Cervigni et al., 2008). In the present work, molecular markers (RAPD, AFLP and SSR) were used on E. curvula genotypes in order to: i) evaluate the usefulness of markers as a methodology for assessing the reproductive mode through progeny tests, ii) compare the efficiency of RAPD markers to assess intracultivar homogeneity or seed purity, iii) establish genetic relationships among the cultivars within the complex, and iv) characterize the new materials obtained in our laboratory.
MATERIALS AND METHODS
Plant Material. Seven apomictic cultivars and a facultative one (cv. Kromdraai) were used. The cultivars, grouped by agronomic type according to Covas (1991), are listed in Table 1. Seeds were either provided by INTA EEA Anguil or obtained from our collection at the Departamento de Agronomía (UNS, Argentina). Seeds were germinated in the greenhouse. Fifteen plants from each accession were planted in pots containing soil and were kept in the greenhouse for two months (winter time, 25 ± 4 °C).
Table 1. Weeping lovegrass materials analyzed, indicating agronomic type (according to Covas 1991), chromosome number and identification code (ID). ¿? Undefined agronomic type.
Tabla 1. Materiales de pasto llorón analizados, indicando el tipo agronómico (de acuerdo a Covas 1991), número de cromosomas y código de identificación (ID). ¿? Tipo agronómico no definido.

The new plant materials obtained by our group as previously reported (Cardone et al., 2006; Mecchia et al., 2006) were also included in this study. They comprised an euploid series obtained from apomictic cv. Tanganyika (2n=4x=40): a diploid sexual line (UNST1122, 2n=2x=20) obtained by in vitro culture and two tetraploid (2n=4x=40) highly sexual plants (UNST1131 and UNST1112) originated from UNST1122 seeds treated with colchicine and DMSO.
In order to characterize the reproductive mode of the plants (apomictic or sexual), progeny tests using RAPD markers were conducted. Progenies consisting of 20-30 individuals, similar to those used in other studies (Daurelio et al., 2004; Matzk et al., 2005), were obtained by open pollination of individuals of apomictic cultivars Morpa and Tanganyika and the sexual genotype UNST1122.
DNA isolation. Genomic DNA was isolated from leaves of two-month-old seedlings grown in the greenhouse. Leaf samples were harvested and homogenized in buffer using mortar and pestle. For approximately 80 mg of leaf tissue 700 µL of extraction buffer was added (50 mM Tris-HCl pH 8.0, 10 mM EDTA pH 8, 100 mM NaCl, 10% (w/v) sodium dodecyl sulfate (SDS) and 10 mM b-mercaptoethanol), and incubated at 65 °C for 20 min. After adding 200 µL of potassium acetate 5 M pH 4.8, the mix was incubated on ice for 20 min, and centrifuged at 13000 rpm for 15 min twice to collect the supernatant. This was followed by precipitation with an equal volume of cold (-20 °C) isopropanol and centrifuged at 13000 rpm for 10 min. The resulting pellet was washed in equal volume of 70% ethanol and centrifuged at 13000 rpm for 4 min twice. Finally, the pellet was air-dried and then dissolved in 50 µL of TE (10 mM Tris-HCl pH 7.6 and 1 mM EDTA) overnight. DNA concentration was determined by fluorochrome spectrophotometry using Hoechst 33258 dye with calf thymus DNA as standard in a BioRad fluorometer. For AFLP assays, genomic DNA was extracted by using a commercial kit (Nucleon PhytoPure, GE Healthcare).
RAPD. Random amplified polymorphic DNA (RAPD) experiments were performed by using the protocol described by the CIMMYT Applied Molecular Genetics Laboratory Protocols, (www.cimmyt.org) with modifications. Five primers (221, 225, 237, 241 and 245) from the NAPS Unit list of standard primers were used. Each amplification reaction was performed in a volume of 25 µL containing 1X Taq polymerase reaction buffer (Invitrogen), 1.5 mM MgCl2, 10 mM each deoxynucleotide triphosphates (dNTPs), 30 ng of each primer, 50 ng of genomic DNA and 1 U of Taq polymerase (Invitrogen). Amplifications were carried out in a MJ Research thermocycler programmed as follows: initial denaturation at 94 °C for 4 min and final extension at 72 °C for 5 min with 36 cycles of 94 °C for 30 sec, 36 °C for 1 min and 72 °C for 1 min. Negative controls without DNA were included. Amplification reliability was assessed by the use of duplicate samples for each genotype. PCR products were mixed with loading dye [98% (v/v) formamide, 10mM EDTA and 0.025% (w/v) bromophenol blue], denatured at 95 °C for 5 min and immediately placed on ice. Reactions were electrophoresed in 6% (w/v) polyacrylamide denaturing gels and silver-stained.
EST-derived simple sequence repeats (EST-SSR). ESTSSR were developed in a previous work (Cervigni et al., 2008) from high quality ESTs using the SSR Discovery program (Robinson et al., 2004). PCR reactions were performed in a final volume of 20 µL containing 1X Taq polymerase reaction buffer (Invitrogen), 2.5 mM MgCl2, 0.125 mM of each dNTPs, 1 µM of each primer, 50 ng of genomic DNA and 2 U of Taq polymerase (Invitrogen). A touchdown program was used consisting of an initial denaturation at 94 °C for 4 min, 15 cycles of 94 °C for 30 sec, 65 °C (-1 °C/cycle) for 1 min and 72 °C for 1 min and 30 cycles of 94 °C for 30 sec, 50 °C for 1 min and 72 °C for 1 min. The final extension step was at 72 °C for 5 min. PCR reactions were performed in an MJ Research thermocycler. Samples were mixed with denaturing loading buffer, treated for 5 min at 95 °C and resolved in 6% (w/v) silver-stained polyacrylamide gels. Thirteen EST-SSR primer combinations that rendered good amplifications were selected for the evaluation of polymorphisms among weeping lovegrass genotypes.
AFLP. AFLP analyses were performed as was described by Vos et al. (1995) with minor modifications. Sequence of adapters, primers containing one selective nucleotide and primers with three nucleotide selective extensions are in Table 2. The preselective and selective amplifications were conducted in a T Gradient thermocycler (Biometra). Final products of the selective PCR reactions were mixed with an equal volume of loading buffer containing 95% (v/v) of formamide including two running dyes (bromophenol blue and xylene cyanol) denatured for 5 min at 95 °C, and immediately chilled on ice. Reactions were electrophoresed in 6% (w/v) polyacrylamide denaturing gels and silver-stained. In order to characterize the genetic variability among weeping lovegrass genotypes, fourteen combinations of AFLP primers +3 were tested.
Table 2. Adapters and primers used for ligation, pre-amplification and amplification reactions of AFLP.
Tabla 2. Adaptadores y primers usados en las reacciones de AFLP para ligación, preamplificación y amplificación.

Data collection and analysis. Each band was considered as an independent locus, and polymorphic bands were scored visually as either absent (0) or present (1) for each one of the 11 genotypes. Only those bands consistently scored were considered for analysis. Differences in band intensity were not considered. Polymorphic bands for each genotype were compiled in a data matrix and analyzed using Numerical Taxonomy and Multivariate Analysis System (NTSYS-pc, Version 2). Jaccard and Simple matching similarity coefficients were calculated for all of the pair-wise comparisons among the genotypes. Cluster analysis was performed according to the unweighted pair group mean algorithm (UPGMA). Data from each class of molecular marker were analyzed independently and then combined in a whole analysis. The correlations between AFLP, SSR and RAPD genetic distance matrices were analyzed by the Mantel test of matrix correspondence. Statistical significance was determined by random permutation, with the number of permutations set to 1000.
RESULTS
Assessment of the reproductive mode. RAPD profiles from progenies coming from individual panicles of the apomictic cultivars Tanganyika and Morpa were uniform (Fig. 1A) while RAPD patterns from offspring of the diploid genotype UNST1122 were variable (Fig. 1B). All the selected RAPD primers assayed on diploid plants rendered polymorphic bands. By combining the markers, unique profiles were obtained for each individual.
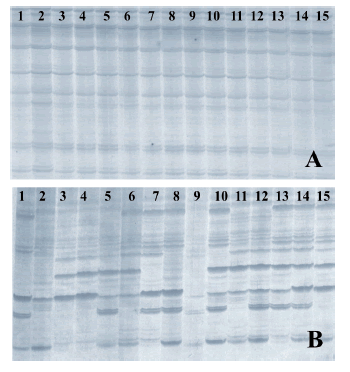
Fig. 1. Progeny test of the apomictic cv. Tanganyika (A) and sexual genotype UNST1122 (B) using RAPD markers in polyacrylamide gels with primers 211 and 222, respectively. Fifteen offspring are numbered.
Fig. 1. Test de progenie del cultivar apomíctico Tanganyika (A) y del genotipo sexual UNST1122 (B) usando marcadores RAPD en geles de poliacrilamida con los primers 211 y 222, respectivamente. Están numerados 15 descendientes.
Intracultivar homogeneity or seed purity. Tests performed by using RAPD markers on plants coming from commercial seeds showed more than one banding pattern in four out of the eight cultivars analyzed. This situation was particularly evident in Morpa, which exhibited six out of 15 off-type individuals (Fig. 2A). Variability was also found within Don Eduardo (four plants) and Tanganyika (one plant) seed sets. Cultivars Don Walter, Don Juan, Ermelo, Don Arturo and Kromdraai were uniform, showing completely monomorphic patterns (Fig. 2B).

Fig. 2. Varietal purity test using RAPD markers on seeds of cvs. Morpa (A), and Don Juan INTA (B). Fifteen plants are numbered.
Fig. 2. Test de pureza varietal usando marcadores RAPD en semillas de pasto llorón cvs. Morpa (A), y Don Juan INTA (B). Están numerados 15 descendientes.
Cultivar differentiation. Variability among cultivars was observed with all the three types of DNA markers assayed, finding 56 polymorphic bands for RAPD, 101 for AFLP and 26 for SSR (Fig. 3). All the primers used for RAPD and AFLP rendered at least one polymorphism among genotypes. Ten out of 13 SSR primers (77%) were polymorphic. Although none of the individual primers was informative enough to identify all the cultivars, combined polymorphisms from all the markers allowed a complete genotype identification.

Fig. 3. Amplification patterns of RAPD (A), SSR (B) and AFLP (C) markers of different genotypes of weeping lovegrass. The letters correspond to genotypes: DW: Don Walter; DJ: Don Juan; DP: Don Pablo; DE: Don Eduardo; K: Kromdraai; DA: Don Arturo; M: Morpa; E: Ermelo; T: Tanganyika; U: UNST1112; V: UNST1122, and B: UNST1131.
Fig. 3. Patrones de amplificación de marcadores RAPD (A), SSR (B) y AFLP (C) de diferentes genotipos de pasto llorón. Las letras corresponden a los genotipos: DW: Don Walter; DJ: Don Juan; DP: Don Pablo; DE: Don Eduardo; K: Kromdraai; DA: Don Arturo; M: Morpa; E: Ermelo; T: Tanganyika; U: UNST1112; V: UNST1122, y B: UNST1131.
The analysis conducted in order to differentiate the agronomic types using AFLP, RAPD and SSR rendered clusters with slight differences. Correlation tests between similarity matrices were high and significant for all the pair comparisons: RAPD vs. AFLP (r =0.81, p<0.001), AFLP vs. SSR (r=0.89, p<0.001) and RAPD vs. SSR (r=0.83, p<0.001). Taking into account the high correlation values, a consensus tree based on data from the three markers was obtained (Fig. 4).

Fig. 4. Consensus tree obtained with the combined data of three molecular markers (AFLP, RAPD, and SSR) of 11 genotypes of weeping lovegrass.
Fig. 4. Árbol consenso obtenido con los datos combinados de los tres marcadores moleculares (AFLP, RAPD y SSR) de los 11 genotipos de pasto llorón.
DISCUSSION
Assessment of the reproductive mode. Uniform RAPD patterns were observed among progenies of cultivars Tanganyika and Morpa, confirming the apomictic nature of these cultivars. The presence of genetic variability among UNST1122 offspring agrees with a sexual reproductive mode, supported by previous cytological evidence (Cardone et al., 2006; Meier et al., 2011). The high level of variability observed in this offspring can be explained by a high heterozygosity level both at mother plant and at several potential pollen donors growing in the same greenhouse, most of them being apomictic polyploid cultivars. Cross-pollination would be guaranteed by a self-incompatibility system, frequently observed at diploid levels in sexual-apomictic species (Bicknell & Koltunow, 2004).
Intracultivar homogeneity or seed purity. Variation in the progeny of an apomictic cultivar could be attributed mainly to three causes: the presence of some level of sexual reproduction, the co-existence of several clones (biotypes) and/or the accidental mixing of seeds during the harvest or storage. Sexual reproduction could be discarded in these cultivars, based on previous studies where they were confirmed as apomictic (Poverene, 1988). Fingerprints of the off-type individuals were analyzed and the abnormal patterns were compared with those of all the other cultivars, in order to explain the intracultivar variation observed in the materials described above. This study showed that three of the abnormal patterns found in Morpa were identical to patterns of Don Walter, two coincided with patterns of Don Eduardo and another one with those found in Don Pablo (robusta type cultivar). Therefore, contamination with seeds from other cultivars seems to be the most plausible explanation for the case of Morpa. With respect to Don Eduardo and Tanganyika, the off-type patterns could not be assigned to any of the other materials included in this analysis; heterogeneity in these cases could be also due to seed mixing with unknown weeping lovegrass materials but an intracultivar co-existence of several clones should not be discarded.
In a previous paper, based on progeny tests, Kromdraai was classified as facultative apomictic, with some plants showing sexual reproduction and others only clonal reproduction (Guzmán et al., 1992). In our study, molecular patterns were completely uniform; suggesting that the analyzed seed pool comes from apomictic plants. In addition to the genotype, many environmental factors, such us photoperiod, temperature or growing conditions might influence the reproductive mode in a facultative apomictic species (Houliston et al., 2006).
Cultivar differentiation. Jaccard and Simple-matching coefficients gave slightly different dendrograms (data not shown). This can be attributed to the inclusion of the negative co-occurrences (band absence) in the simple-matching coefficient. This does not mean that the DNA regions considered are identical (da Silva Meyer et al., 2004). For that reason the Jaccard similarity matrix was chosen to perform cluster analysis.
In the three dendrograms obtained with AFLP, RAPD and SSR, the curvula-type cultivars formed a clear defined group, being Don Walter (conferta) and UNST1122 the closest ones to the curvulas. The rest of the agronomic types are distributed in the tree within variable similarity coefficients. A close relationship between the conferta and curvula types was previously observed and a common origin for these strains has been proposed. Conferta lovegrass could be one of the ancestral parents of the weeping form and a founder of the E. curvula complex (Poverene & Voigt, 1997). Clusters obtained using RAPD and AFLP were similar to each other and showed concordance with previous studies based on isozyme markers (Poverene, 1988; Poverene& Voigt, 1997).
AFLP were the most informative markers since they allowed the differentiation among all the commercial cultivars. Cultivars Ermelo, Morpa and Don Arturo remained indistinguishable by RAPD and SSR markers. The reason of these identities could be found in the origin of these cultivars. The variety protection certificate describes Morpa as a derivative cultivar (USDA, 1972). Poverene (1988) suggests that Ermelo and Morpa were selected from the same ecotype and Don Arturo is also believed to have been so selected. Neither morphological traits nor seed isozymes could differentiate one cultivar from the other (Poverene, 1988). Besides the high morphological and molecular similarity, selection has probably increased the palatability of Morpa, based on meat production information (Voigt et al., 1970).
Regarding the new genotypes obtained by tissue culture and colchicine treatment, the tetraploids UNST1131 and UNST1112 grouped together within the curvula type and closely linked to Tanganyika, which is the cultivar from which they were originated. AFLP and RAPD allowed the differentiation of these genotypes from Tanganyika and only with RAPD it was possible to differentiate between UNST1131 and UNST1112.
The diploid genotype UNST1122 showed to be more similar to the conferta type (Don Walter) based on two types of molecular markers. Moreover, this genotype is morphologically similar to cultivars of the robusta type and shows high vigor (Cardone et al., 2006). These are unexpected results considering that it derives from Tanganyika, a curvula type cultivar. Similar results were observed by Voigt et al. (1972) and Poverene and Voigt (1997) for another diploid material (PI299920) using morphological traits and isozymes. These authors found the accession PI299920 to be more similar to robusta than to the curvula type, although it was not well linked to any of the defined agronomic types. For this reason the authors supposed that, rather than being an ancestral diploid, PI299920 was derived from higher ploidy levels (Voigt et al., 2004). Similar observations were made by Stupar et al. (2007) in a potato euploid series, where artificial diploids were more vigorous than the original tetraploids. Ploidy is an important factor that confers special and typical features to each level, also in E. curvula (Mecchia et al., 2007). Based on the mentioned differences we proposed to create a new type to include the diploids of E. curvula.
Jaccard similarity coefficients ranged from 0.31 to 1. Genetic similarity measures among cultivars can vary within Eragrostis species. In previous reports similarity values for E. tef accessions fell within the range of 0.84-0.96 with RAPD markers (Bai et al., 2000), indicating scarce variability at DNA level that might be due to a narrow genetic base in the species. The occurrence of self-pollination in E. tef could even decrease the genetic variation within the species. It is known that accessions of weeping lovegrass come from not too many materials collected in Southern Africa and then distributed in US and Argentina. However, the variability level observed shows that the materials introduced into our country retained significant genetic diversity, which represents an excellent perspective for improvement.
The reproductive mode of weeping lovegrass (apomixis) seems to have played a fundamental role in producing a complex superstructure of polyploid forms, as has happened with other grasses (Quarín & Burson, 1991; Adamowski et al., 2005). While sexuality, through the fusion of unreduced gametes, would allow the emergence of polyploid types that are progressively more complex, apomixis would ensure the survival of these meiotically unstable new lines. The E. curvula complex consists of a set of populations, most of which are clonal with a smaller proportion exhibiting sexual reproduction. Crosses between both populations continuously add new genotypes to the complex, which in this way constitutes a true collective species (Poverene, 1988). The net result is a labile, dynamic and highly adaptable system capable of a high degree of adaptive polymorphism and rapid evolution (Harlan & de Wet, 1963).
SSR analysis was less informative in characterizing closely related weeping lovegrass genotypes, compared with RAPD and AFLP. An explanation for these results could be that repetitive microsatellites motifs were developed by searching in four ESTs databases of E. curvula (Cervigni et al., 2008). EST-SSR has been reported to be less polymorphic compared with genomic SSR because of greater DNA sequence conservation in transcribed regions (Varshney et al., 2005). EST-SSR has some intrinsic advantages over genomic SSR because they are quickly obtained by electronic sorting, and are present in expressed regions of the genome. The usefulness of these markers also lies in their expected transferability because the primers are designed from the more conserved coding regions of the genome (Varshney et al., 2005). SSR markers derived from genes produce a high proportion of high-quality markers with strong bands and distinct allelic peaks in most reports (Thiel et al., 2003). In a previous study (Cervigni et al., 2008), sixty EST-SSR primers were analyzed over weeping lovegrass DNA and 53% of them rendered successful amplification. Saha et al. (2004) reported that 92% of the tall fescue-derived ESTSSR primer pairs produced characteristic SSR bands and approximately half of them were polymorphic between parents of tall fescue and ryegrass populations, as well as in rice and wheat.
Microsatellites met the four criteria proposed by Bailey (1983) for an optimal marker: 1) maximum variation between cultivars, 2) minimal intracultivar variation; 3) environmental stability and 4) experimental reproducibility. In our study, SSR were the less informative markers because they did not render a complete cultivar identification. However, clusters based on SSR showed a high resemblance to the previous ones based on isozymes and morphological traits (Poverene & Voigt, 1997; Covas, 1991). A lower resolution in our study indicates that it would be necessary to increase the number of primers in order to resolve all the genotypes.
At the moment, there is no available information related to the identification of weeping lovegrass accessions at the DNA level. Molecular fingerprinting will be helpful in assessing the purity and stability of genotypes entering into a breeding and/or seed multiplication program, and in reducing duplicate germplasm collections. These utilities maximize the genetic diversity, minimize the cost of maintaining germplasm, and eliminate environmental variation, which is unavoidable when morphological markers are used.
From the above we conclude:
Molecular markers are very useful tools to assess, through progeny tests, the reproductive mode of weeping lovegrass plants.
It is possible to determine the intracultivar homogeneity or seed purity in weeping lovegrass and to detect the presence of contamination with seeds from other sources.
AFLP are the markers with the highest potential for cultivar discrimination. The clustering using SSR and AFLP were consistent with previous studies carried out with isozymes and morphological traits.
In the light of these results the new materials developed in our laboratory should be classified as curvula type in the case of UNST1131 and UNST1112. For UNST1122, the creation of a new morphological type is proposed because it posses morphological characteristics of robusta and molecular profiling of conferta type.
ACKNOWLEDGEMENTS
The project was granted by the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT), grants PAV 137/4 and PICT 14625, the Secretaría de Ciencia y Tecnología (SECyT UNS) grant PGI 24/A133 (Argentina) and CONICET (PIP 112-200801-01517).
REFERENCES
1. Ayele, M., J. Dolezel, M. Van Duren, H. Brunner & F. Zapata-Arias (1996). Flow cytometric analysis of nuclear genome of the Ethiopian cereal tef [Eragrostis tef (Zucc.) Trotter]. Genetica 98: 211-215. [ Links ]
2. Bai, G., M. Ayele, H. Tefera & H. Nguyen (2000). Genetic diversity in tef and its relatives as revealed by RAPDs. Euphytica 112: 15-22. [ Links ]
3. Bailey, D. (1983). Isozymic variation and plant breeder´s rights. In: Tanksley, S. and J. Orton (eds). Isozymes in plant genetics and breeding. Part A. Elsevier, Amsterdam, pp. 425-441. [ Links ]
4. Bennett, M. & J. Smith (1976). Nuclear DNA amounts in angiosperms. Philosophical Transactions of the Royal Society of London, Series B 274: 227-274. [ Links ]
5. Bicknell, R. & A. Koltunow (2004). Understanding apomixis: recent advances and remaining conundrums. Plant Cell 16: S228-S245. [ Links ]
6. Cardone, S., P. Polci, J.P. Selva, M. Mecchia, S. Pessino, P. Hermann, V. Cambi, P. Voigt, G. Spangenberg & V. Echenique (2006). Novel genotypes of the subtropical grass Eragrostis curvula for the study of apomixis (diplospory). Euphytica 151: 263-272. [ Links ]
7. Cervigni, G., N. Paniego, M. Diaz, J.P. Selva, D. Zappacosta, D. Zanazzi, I. Landerreche, S. Felitti, S. Pessino, G. Spangenberg& V. Echenique (2008). Expressed sequence tag analysis and development of gene associated markers in a near-isogenic plant system of Eragrostis curvula. Plant Molecular Biology 67: 1-10. [ Links ]
8. Covas, G. (1991). Taxonomía y morfología del pasto llorón [Eragrostis curvula (Schrad.), Nees], con referencias sobre otras especies cultivadas de Eragrostis. In: Fernández, O., R. Brevedan & A. Gargano (eds). El pasto llorón, su biología y manejo. CERZOS and Universidad Nacional del Sur, Bahía Blanca, Argentina, pp. 7-17. [ Links ]
9. da Silva Meyer, A., A. Franco García, A. Pereira de Souza & C. Lopes de Souza (2004). Comparison of similarity coefficients used for cluster analysis with dominant markers in maize (Zea mays L). Genetics and Molecular Biology 27: 83-91. [ Links ]
10. de Winter, B. (1995). Eragrostis Beauv. The grasses and pastures of South Africa. Central News Agency. Union of South Africa, p. 132-184. [ Links ]
11. Guzmán, N., M. Di Renzo & M. Poverene (1992). Determinación de la variación isoenzimática en progenies de Eragrostis curvula (Schrad.) Nees. Turrialba 42: 321-326. [ Links ]
12. Harlan, J. & J. de Wet (1963). The role of apomixis in the evolution of the Bothriochloa-Dichanthium complex. Crop Science 3: 314-316. [ Links ]
13. Houliston, G., H. Chapman & R. Bicknell (2006). The influence of genotype and environment on the fecundity and facultative expression of apomixis in Hieracium pilosella. Folia Geobotanica 41: 165-181. [ Links ]
14. Jaccard, P. (1908). Nouvelles recherches sur la distribution florale. Bulletin Société Vaudoise des Sciences Naturelles 44: 223-270. [ Links ]
15. Jacobs, S. (1982). Classification in the Eragrostis curvula complex in Australia. Australian Plant Introduction Review CSIRO 15: 5-14. [ Links ]
16. Leigh, J. (1961). Leaf anatomy in certain strains of Eragrostis (Beauv.). Journal of South African Botany 27: 141-146 [ Links ]
17. Leigh, J. & R. Davidson (1968). Eragrostis curvula (Schrad.) Nees and some other African lovegrasses. Australian Plant Introduction Review CSIRO 5: 21-44. [ Links ]
18. Mecchia, M., A. Ochogavıa, J. P. Selva, N. Laspina, S. Felitti, L. Martelotto, G. Spangenberg, V. Echenique & S. Pessino (2007). Genome polymorphisms and gene differential expression in a 'back-and-forth' ploidy-altered series of weeping lovegrass (Eragrostis curvula). Journal of Plant Physiology 164: 1051-1061. [ Links ]
19. Meier, M., D. Zappacosta, J.P. Selva, S. Pessino & V. Echenique. (2011). Evaluation of different methods for assessing the reproductive mode of weeping lovegrass plants, Eragrostis curvula (Schrad.) Nees. Australian Journal of Botany 59: 253-261. [ Links ]
20. Poverene, M. (1988). Contribución citogenética y quimiosistemática a la taxonomía del pasto llorón, Eragrostis curvula (Schrad.) Nees. Doctoral Thesis. Universidad Nacional del Sur, Argentina. [ Links ]
21. Poverene, M. & P. Voigt (1997). Isozyme variation and germplasm relationships in the Eragrostis curvula complex. Biochemical Systematics and Ecology 25: 21-32. [ Links ]
22. Quarín, C., M. Pozzobon & J. Valls (1996). Cytology and reproductive of diploid, tetraploid and hexaploid germplasm accessions of a wild forage grass: Paspalum compressifolium. Euphytica 90: 345-349. [ Links ]
23. Robinson, A., G. Christopher, C. Love, J. Batley, G. Barker & D. Edwards (2004). Simple sequence repeat marker loci discovery using SSR primer. Bioinformatics 20: 1475-1476. [ Links ]
24. Roodt-Wilding, R. & J. Spies (2006). Phylogenetic relationships in southern African chloridoid grasses (Poaceae) based on nuclear and chloroplast sequence data. Systematics and Biodiversity 44: 401-415. [ Links ]
25. Saha, M., M. Mian, I. Eujayl, J. Zwonitzer, L. Wang & G. May (2004). Tall fescue EST-SSR markers with transferability across several grass species. Theoretical and Applied Genetics 109: 783-791. [ Links ]
26. Stupar, R., P. Bhaskar, B. Yandell, W. Rensink, A. Hart, S. Ouyang, R. Veilleux, J. Busse, R. Erhardt, C. Buell & J. Jiang (2007). Phenotypic and transcriptomic changes associated with potato autopolyploidization. Genetics 176: 2055-2067. [ Links ]
27. Thiel, T., W. Michalek, R. Varshney & A. Graner (2003). Exploiting EST databases for the development and characterization of genederived SSR-markers in barley (Hordeum vulgare L.). Theoretical and Applied Genetics 106: 411-422. [ Links ]
28. Varshney, R., A. Graner & M. Sorrells (2005). Genic microsatellite markers in plants: features and applications. Trends in Biotechnology 23: 48-55. [ Links ]
29. Voigt, P. (1970). New varieties of weeping lovegrass through plant evaluation and selection. Proceedings of the first weeping lovegrass symposium, The Samuel Roberts Noble Foundation, Ardmore, USA, April 28 and 29, pp. 14-20. [ Links ]
30. Voigt, P. & E. Bashaw (1972). Apomixis and sexuality in Eragrostis curvula. Crop Science 12: 843-847. [ Links ]
31. Voigt, P., N. Rethman & M. Poverene (2004). Lovegrasses. In: Moser, L. (ed.). Warm-Season (C4) Grasses, Agronomy Monograph 45, American Society of Agronomy. Madison WI, USA, [ Links ]
32. Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman & M. Kuiper (1995). AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research 23: 4407-4414. [ Links ]
33. Yu, J., E. Graznak, F. Breseghello, H. Tefera & M. Sorrells (2007). QTL mapping of agronomic traits in tef. BMC Plant Biology 7: 30-42. [ Links ]














