Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
Phyton (Buenos Aires)
versión On-line ISSN 1851-5657
Phyton (B. Aires) vol.81 no.1 Vicente López ene./jun. 2012
ORIGINAL ARTICLES
Construction and characterization of a metagenomic DNA library from the rhizosphere of wheat (Triticum aestivum)
Construcción y caracterización de una biblioteca de ADN metagenómico de la rizósfera de trigo (Triticum aestivum)
Hernández-León R1, M Martínez-Trujillo2, E Valencia-Cantero1, G Santoyo1
1 Instituto de Investigaciones Químico Biológicas. Universidad Michoacana de San Nicolás de Hidalgo. Morelia, Michoacán, México.
2 Facultad de Biología, Universidad Michoacana de San Nicolás de Hidalgo. Morelia, Michoacán, México.
Address Correspondence to: Dr. Gustavo Santoyo. Instituto de Investigaciones Químico Biológicas. Universidad Michoacana de San Nicolás de Hidalgo. Ciudad Universitaria edif. A1', 58030. Morelia, Michoacán, México, e-mail: gustavo_santoyo@yahoo.com; gsantoyo@umich.mx
Recibido / Received 3.IX.2011.
Aceptado / Accepted 14.XI.2011.
Abstract. Rhizospheric soil of wheat plants contains a high diversity of microorganisms, and therefore, comprises a large reservoir for discovering genes with diverse agro-biotechnological applications. In this work, we constructed an E. coli metagenomic library based on bacterial artificial chromosome (BAC) clones with large genomic inserts from metagenomic DNA from the rhizosphere of wheat plants. The average of the DNA cloned segments varies from 5 to 80 kb, with an average size of 38 kb. Random clones were end-sequenced and homology results showed that the clonation of metagenomic DNA codes mainly for metabolic and catalytic functions (40%), including amidohydrolase, hydrolase, peptidase, serine protease, endonuclease and exonuclease. Another interesting proportion of the clones revealed genomic sequences with hypothetical (17%) or unknown function (9%). The metagenomic sequences belonged mostly to Proteobacteria, Firmicutes, Archaea, Actinobacteria, Fungi, Virus, Unclassified Bacteria and Unknown taxa. It is expected that the metagenomic library from rhizospheric soil of wheat plants keeps growing. At the same time, functional analyses are conducted in the search for genes of agrobiotech interest.
Keywords: Metagenomics; Rhizosphere; Wheat.
Resumen. El suelo rizosférico de las plantas de trigo contiene una alta diversidad de microorganismos y por lo tanto, constituye una gran reserva para el descubrimiento de los genes con diversas aplicaciones de la biotecnología agrícola. En este trabajo, hemos construido una biblioteca metagenómica empleando como huésped heterólogo E. coli, clonando grandes fragmentos de ADN genómico en un cromosoma artificial bacteriano (BAC), proveniente de la rizósfera de plantas de trigo. El promedio de los segmentos de ADN clonado varió de 5 a 80 kb, con un tamaño promedio de 38 kb. Los extremos de varias clonas BAC, tomadas al azar fueron secuenciadas. Los resultados de homología de los BACs analizados mostraron principalmente funciones metabólicas y catalíticas (40%), incluyendo amidohidrolasas, hidrolasas, peptidasas, serina proteasas, endonucleasas y exonucleasas. Otra parte interesante de las clonas reveló secuencias genómicas con función hipotética (17%) o desconocida (9%). Las secuencias metagenómicas pertenecen en su mayoría a Firmicutes, Proteobacterias, Arqueas, Actinobacterias, hongos, virus, bacterias y taxones desconocidos sin clasificar. Se espera que la biblioteca metagenómica del suelo rizosférico de las plantas de trigo siga creciendo. Al mismo tiempo, se están llevando a cabo análisis funcionales en la búsqueda de genes de interés agro-biotecnológico.
Palabras clave: Metagenómica; Rizósfera; Trigo.
INTRODUCTION
Metagenomics has revolutionized the way people study biology. In fact, there are currently more metagenomic sequences reported in databases such as NCBI than the total sequences of all completely sequenced organisms. This represents a challenge to biology (Moreno-Hagelsieb & Santoyo, unpublished results). Metagenomics has been investigated from various environments, including gut microbiota, aquatic ecosystems, mines, agricultural and forest soils (Kim et al., 1996; Rondon et al., 2000; Wang et al., 2000; Lee et al., 2004; Tyson et al., 2004; Craig et al., 2009; Qin et al., 2010). The relevant insight of these studies is that each of them has shown different aspects to study and analyze. In some cases, they have provided innovative views of the microbial ecology on extreme environments for the development of life. At the same time, others have found novel genetic elements that could have applications in biotech industry (Rondon et al., 2000; Wang et al., 2000; Tyson et al., 2004; Nacke et al., 2011).
One of the most complex environments is the rhizosphere, a microenvironment where a great microbial diversity is present. The rhizosphere is defined as the region of soil which is influenced by the plant roots (Ahmad et al., 2008). It is in this environment where various abiotic and biotic interactions take place, especially concerning microorganisms and plant roots. Abiotic interactions, such as temperature, soil type and climate (among other factors) are often involved in determining the population structure of microbial communities. Microorganisms of these communities, however, have evolved mechanisms to deal with occupation of the space which allows them to obtain nutrients excreted by the plant roots (Weller, 1988; Haas & Keel, 2003). The organism diversity of the bulk-soil or the rhizospheric soil of different plants has been extensively studied; most studies have reported a wide range of organisms from those soil portions (Daniel, 2005). It is estimated that there are about 2x106 bacterial species in marine environments. Surprisingly, a soil sample could contain up to 4x106 different taxa (Curtis et al., 2002). This suggests that soils are a large reservoir for the discovery of several compounds that may have applications in agriculture, human health or industry (Handelsman, 2004; Hernandez-Leon et al., 2010). In this work, we report the construction of a bacterial artificial chromosome (BAC) metagenomic DNA library from the rhizosphere of wheat plants. We also characterized and sequenced various clones that contain genes with different catalytic activities, and sequences from different phyla (including non-cultivable or even unknown microorganisms).
MATERIALS AND METHODS
Sampling and physico-chemical analysis of rhizospheric soil. To study the soil microbial community, samples were taken from the rhizosphere of wheat plants near the city of Zamora, Michoacan, Mexico (19° 59' N, 102° 17' W, 1560 m.a.s.l.). Ten plants were collected along with their roots and rhizospheric soil, one month after planting. Rhizospheric soil samples were taken at 10 cm depth from the soil surface, and transported on ice to be stored at 4 °C for immediate analysis in the laboratory. The rhizosphere was separated from the roots, and stored at -4 °C.
The soil physico-chemical properties were as follows: the soil was a clay loam type, with 23.48% sand, 38.57% clay and 38% silt. The pH was neutral (7.17). Organic matter was 3.73%, inorganic nitrogen 16.9 ppm, potassium 653 ppm, calcium 4938 ppm, magnesium 1416 ppm, sodium 153 ppm, iron 22.8 ppm, zinc 0.91 ppm, manganese 55.9 ppm and copper 2.55 ppm. These physico-chemical conditions resulted optimal for cropping wheat.
DNA extraction and purification. Total DNA extraction from the rhizospheric soil samples was done using the MO-BIO PowerSoil® DNA Isolation Kit, following manufacturer instructions. Extracted DNA solution was completely transparent, checked by gel electrophoresis and stained with ethidium bromide (4 µg/mL).
The extracted DNA was purified using a Wizard SV Genomic DNA minicolumn purification System from Promega. The integrity of DNA was checked by gel electrophoresis (at 1%, Fig.1a). DNA was tested using small quantities of metagenomic DNA as a template for PCR amplification of the 16S ribosomal genes with the universal primers FD1 and RD1: 5´-CAG AGT TTG ATC CTG GCT CAG-3´and 5´-AAG GAG GTG ATC CAG CC-3´ (Weisburg et al., 1991; Velazquez-Sepulveda et al., 2011). Once DNA was pure enough, approximately 300 ng of DNA were digested with the HindIII enzyme for 4 hr at 37 °C. The digestion was again observed in a gel electrophoresis at 1%. Then, the restriction enzyme was inactivated at 65 °C in a Dry Bath Incubator (Fisher Scientific) during 20 min, and purified by minicolumn (Wizard SV Genomic DNA Purification System, Promega). Thereafter, it was observed in an agarose gel at 1% (Fig 1c and d).

Fig. 1. (a) Metagenomic DNA from the rhizosphere of wheat plants; (b) molecular size of a DNA marker; (c, d) metagenomic DNA partially digested with HindIII.
Fig. 1. (a) DNA metagenómico de la rizósfera de plantas de trigo; (b) tamaño molecular de un marcador de ADN; (c, d) DNA metagenómico parcialmente digerido por HindIII.
Metagenomic library construction. To perform the cloning of metagenomic DNA, 50 ng were used plus 10 ng of the vector pIndigoBAC-5, incubating with the T4 ligase enzyme (4 units) during 12 hrs at 4 °C. The ligation product was transformed by electroporation into electrocompetent E. coli cells in a 2510 Eppendorf® electroporator. The transformed cells were plated onto LB solid medium with chloramphenicol (Cm, 10 µg/mL) and incubated for 20 to 24 hrs at 37 °C. Clones were picked out again on LB medium plates containing 10 µg/mL of Cm plus 40 µL of X-gal (at a concentration of 60 mg/mL) and incubated for 24 hrs at 37 °C.
RESULTS
Insert size analysis of the library. The construction of metagenomic libraries from different environments has shown a great potential to identify new or improved genetic determinants that can be used in different areas of medicine, industry and agrobiotechnology (Van Elsas, 2008). DNA from different types of soil makes things difficult due to the high content of soil components that inhibit Taq polymerases and restriction enzymes, among others. Therefore, obtaining high quality and purified DNA from this environment is essential, especially for future cloning steps. Figure 1a shows the isolation of metagenomic DNA with excelent quality, including second steps for purification and enzyme digestion with HindIII in agarose gel (Fig. 1c and d). Control experiments to determine the purity of metagenomic DNA showed high quality, even managed to be used as a template for 16S ribosomal gene amplification by PCR (data not shown). As can be seen, the digestion was partial and obtained a DNA large fragments (> 30 kb), which is recommended for the construction of libraries based on BACs (Handelsman, 2004).
Metagenomic libraries can be divided into two classes: those with small or large inserts. We report the latter class in this work. We isolated at random BACs of 26 recombinant E. coli clones and digested with HindIII to determine the approximate size of the cloned fragments. Figure 2 shows that the cloned inserts in the clones analyzed ranged from 5 to over approximately 80 kb, with an average of 38 kb in size of the inserts. This range is essential to carry out the cloning of genes or operons that may encode complete new metabolic functions (Daniel et al., 2005).
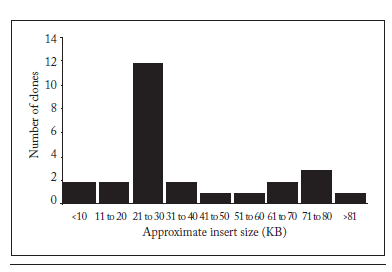
Fig. 2. Approximate metagenomic DNA insert size of 26 BAC clones. See text for details.
Fig. 2. Tamaño aproximado de la inserción de ADN metagenómico de 26 clones BAC. Ver el texto por detalles.
Construction and analysis of the metagenomic library. To get a better idea of what kind of genes or sequences were cloned in the metagenomic library, 75 recombinant clones were isolated and BAC-end commercially sequenced. Twenty three percent of the sequences coded for metabolic functions, while another large percentage corresponded to catalytic functions (17%). Genes encoding amidohydrolase, hydrolase, peptidase, serine protease, endonuclease and exonuclease activities were found, among others (Fig. 3). Interestingly, a significant percentage identity was not found with known sequences or genes are reported as hypothetical, indicating that they might encode for functions still unknown. Moreover, identity was obtained for genes involved in basic cellular processes such as replication, transcription, translation and repair of DNA, as well as genes with identity to membrane proteins and transposons (Fig. 3). The sequences found in the BACs correspond mainly to Alpha and Gammaproteobacteria, Fimicutes, Archaea and Betaproteobacteria (Fig. 4). Other sequences showed identity with fungi, viruses and Cyabonacteria, representing a wide diversity of organisms in the rhizosphere of wheat plants.

Fig. 3. Predicted functional activities of the sequenced DNA inserts of the metagenomic library. See text for details.
Fig. 3. Actividades funcionales predichas de las inserciones de ADN secuenciado de la biblioteca metagenómica. Ver el texto por detalles.

Fig. 4. Predicted taxonomic association of the sequenced DNA inserts of the BAC clones. See text for details.
Fig. 4. Asociación taxonómica predicha por las inserciones de ADN secuenciado de los clones BAC. Ver el texto por detalles.
DISCUSSION
In this work we reported the construction of a large insert size metagenomic DNA library from the rhizosphere of wheat plants. Large inserts are important to isolate complete genes or even complete operons encoding functional pathways for synthesis of diverse compounds or metabolites. In this way, Chung et al. (2008) reported the cloning of a fragment of 40 kb DNA from forest soil metagenomic containing a complete operon encoding type II familypolyketide synthases, ACP synthases, aminotransferase and an ACP reductase, which showed antifungal activity. Nonribosomal peptide synthetases and polyketide synthases are multiprotein complexes (which are encoded by clusters of genes), transcriptional units usually involving operons with several kilobases of length. Other metabolites, such as siderophores, which are big peptide molecules with affinity for iron, are also synthesized by some nonribosomal peptide synthetases. Siderophores are important molecules to colonize and deprive iron to pathogens from rhizospheric soils, thus inhibiting their growth in an indirect form (Rondon et al., 2004). Other compounds involved in the inhibition of pathogens such as lipopeptides from Pseudomonas and Bacillus species, also require clusters of genes for its synthesis. Therefore, it is desirable to clone large DNA if anyone wants to find antifungal activities from bulk-soils or rhizosphere soils. Likewise, Daniel in 2005 suggested that the construction of metagenomic libraries of large inserts has several advantages for detecting genes with desirable functions, such as requiring a small number of clones for screening, and finding positive outcomes. However it can also have some disadvantages, mainly technical, because of the difficulty of obtaining DNA of good purity and cloning of large DNA fragments. Once you can solve these problems, it has obvious advantages over the construction of large rather than small insert libraries.
The rhizosphere of plants is one of the most studied ecosystems. However, in our library 4% of the sequences showed no identity with known sequences in NCBI databases, which represents a significant potential for discovering new functions or unknown organisms in environments widely studied (Fig. 3). The clonation of environmental DNA and expression in BAC vectors is an interesting option to study the metagenome of an ecosystem. In addition, it does not require or rely on culture methods that limit their study to those organisms that can only be reproduced in laboratory. Several microorganisms have been used as hosts for expression of heterologous genes, including Ralstonia metallidurans, Streptomyces lividans, S. cerevisiae and E. coli (Rondon et al., 2000; Chung et al., 2008; Craig et al., 2009). However, each host has constraints, and is able to express genes that come from other organisms. However, E. coli appears to be the Trojan Horse to carry out genes from many different phyla (Handelsman, 2004; Hernández-León et al., 2010). It would be interesting to analyze the metagenomic DNA from the soil in other hosts. This is because a large proportion of bacteria (e.g.: the genus Bacillus) can represent an important part of the microorganisms found in bulk- or rhizospheric soils. This would represent the possibility of cloning other genes which are strictly expressed in Gram-positive bacteria.
In a bulk-soil library from Wisconsin, it was reported the cloning and expression of genes coding for lipase, amylase, hemolytic activity, antibacterial and nucleases (Rondon et al., 2000). Also, some 16S rDNA genes were isolated and sequenced from the same library, including sequences belonging to different microbial phyla, such as low-G + C Grampositive Acidobacterium, Cytophagales, and Proteobacteria. In our library, we report the isolation of 16S ribosomal genes from metagenomic DNA directly (Velazquez-Sepulveda et al., 2012), and not from cloned inserts in the library; here, we found a variety of ribosomal sequences belonging to Classes Alfaproteobacteria, Betaproteobacteria, Deltaproteobacteria, Gammaproteobateria, Actinobacteria, Bacilli, Clostridia bacteria and Unculturable. Within the class Gammaproteobacteria, the genus Pseudomonas, Bacillus and Stenotrophomonas were the most abundant, since they corresponded to 40% of the whole ribosomal library. Previous work also suggested that in principle there was a large bacterial diversity in this environment: Shannon-Wiener test indicated a rate of 3.8 bits per individual (Velazquez-Sepulveda et al., 2012). In this work, our results of sequence analysis in 75 BACs show that inserts mainly belong to Proteobacteria, Firmicutes and Archaea, which coincides with the analysis of bacterial diversity isolating and sequencing ribosomal sequences (Velazquez- Sepulveda et al., 2012).
Metagenomics, and especially soil metagenomics promises to discover new or improved molecules that may have several biotechnological applications. However, this complex ecosystem is considered particularly a reservoir of genetic material that encodes for molecules with some activity in biocontrol (or even direct plant-growth promoting characteristics). Such is the case of international efforts conducted under the project Metacontrol, which aims to analyze various metagenomic DNAs from disease-suppressive soils (Van Elsas et al., 2008). Therefore, the pursuit of such activities is demonstrated in the library reported in this paper, as well as ongoing genomic cloning of more genetic material from the metagenome of wheat rhizospheric soils from crops of Mexico, which represents a relevant option to study due to its high microdiversity.
ACKNOWLEDGEMENTS
We thank Consejo Nacional de Ciencia y Tecnología, México (Proyecto No. 169346) and Coordinación de la Investigación Científica-Universidad Michoacana de San Nicolás de Hidalgo (2011-2012) for financial support to this project. We thank M. Govindappa and reviewers for their comments and suggestions to improve the paper.
REFERENCES
1. Ahmad, F., I. Ahmad, & M.S. Khan (2008). Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Research in Microbiology 163: 173-81. [ Links ]
2. Chung, E.J., H.K. Lim, J.C. Kim, G.J. Choi, E.J. Park, M.H. Lee, Y.R. Chung & S.W. Lee (2008). Forest soil metagenome gene cluster involved in antifungal activity expression in Escherichia coli. Applied and Environmental Microbiology 74: 723-30. [ Links ]
3. Craig, J.W., F.Y. Chang & S.F. Brady (2009). Natural products from environmental DNA hosted in Ralstonia metallidurans. ACS Chemical Biology 4: 23-28. [ Links ]
4. Curtis, T.P., W.T. Sloan & J.W. Scannell (2002). Estimating prokaryotic diversity and its limits. Proceedings of the National Academy of Sciences USA 99: 10494-10499. [ Links ]
5. Daniel, R. (2005). The metagenomics of soil. Nature Review Microbiology 3: 470-478. [ Links ]
6. Haas, D. & C. Keel (2003). Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annual Review of Phytopathology 41: 117-53. [ Links ]
7. Hernández-Leon, R., I. Velazquez-Sepulveda, M.C. Orozco- Mosqueda & G. Santoyo (2010). Metagenómica de suelos: grandes retos y nuevas oportunidades biotecnológicas. Phyton, International Journal of Experimental Botany 79: 4-11. [ Links ]
8. Kim, U.-J., B. B. Birren, T. Slepak, V. Mancino, C. Boysen, H.-L. Kang, M. I. Simon & H. Shizuya (1996). Construction and characterization of a human bacterial artificial chromosome library. Genomics 34: 213-218. [ Links ]
9. Lee, S.W., K. Won, H.K. Lim, J.C. Kim, G.J. Choi & K.Y. Cho (2004). Screening for novel lipolytic enzymes from uncultured soil microorganisms. Applied Microbiology and Biotechnology 65: 720-726. [ Links ]
10. Nacke, H., C. Will, S. Herzog, B. Nowka, M. Engelhaupt & R. Daniel (2011). Identification of novel lipolytic genes and gene families by screening of metagenomic libraries derived from soil samples of the German Biodiversity Exploratories. FEMS Microbiology Ecology doi: 78:188-201. [ Links ]
11. Qin, J., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59-65. [ Links ]
12. Rondon, M.R., P.R. August, A.D. Bettermann, S.F. Brady, T.H. Grossman, M.R. Liles, K.A. Loiacono, B.A. Lynch, I.A. MacNeil, C. Minor. C.L. Tiong, M. Gilman, M.S. Osburne, J. Clardy, J. Handelsman & R.M. Goodman (2000). Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Applied Environmental Microbiology 66: 2541-2547. [ Links ]
13. Rondon, M.R., K.S. Ballering & M.G. Thomas (2004). Identification and analysis of a siderophore biosynthetic gene cluster from Agrobacterium tumefaciens C58. Microbiology 150: 3857-66. [ Links ]
14. Tyson, G.W., J. Chapman, P. Hugenholtz, E.E. Allen, R.J. Ram, P.M. Richardson, V.V. Solovyev, E.M. Rubin, D.S. Rokhsar & J.F. Banfield (2004). Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428: 37-43. [ Links ]
15. van Elsas, J.D., R. Costa, J. Jansson, S. Sjöling, M. Bailey, R. Nalin, T.M. Vogel & L. van Overbeek (2008). The metagenomics of disease-suppressive soils - experiences from the METACONTROL project. Trends in Biotechnology 26: 591-601. [ Links ]
16. Velazquez-Sepulveda, I., M.C. Orozco-Mosqueda, C.M. Prieto- Barajas & G. Santoyo (2012). Bacterial diversity associated with the rhizosphere of wheat plants (Triticum aestivum): Toward a metagenomic analysis. Phyton, International Journal of Experimental Botany 81: 81-87. [ Links ]
17. Wang, G.Y., E. Graziani, B. Waters, W. Pan, X. Li, J. McDermott, G. Meurer, G. Saxena, R.J. Andersen & J. Davies (2000). Novel natural products from soil DNA libraries in a streptomycete host. Organic Letters 2: 2401-2404. [ Links ]
18. Weisburg, W.G., S.M. Barns, D. A. Pelletier & D. J. Lanet (1991). 16S Ribosomal DNA Amplification for Phylogenetic Study. Journal of Bacteriology 173: 697-703. [ Links ]
19. Weller, D.M. (1988). Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annual Review of Phytopathology 26: 379-407. [ Links ]














