Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
Phyton (Buenos Aires)
versión On-line ISSN 1851-5657
Phyton (B. Aires) vol.81 no.1 Vicente López ene./jun. 2012
SHORT NOTE
Chromosomal location of four genes encoding Class III peroxidases in wheat
Localización cromosómica de cuatro genes de peroxidasa de Clase III en trigo
Simonetti E1, E Alba2, A Delibes2
1 Cátedra de Microbiología Agrícola/INBA (CONICET/UBA), Facultad de Agronomía, Universidad de Buenos Aires. Av. San Martín 4453 C1417DSE, Buenos Aires, Argentina.
2 Departamento de Biotecnología, ETS Ing. Agrónomos, Universidad Politécnica de Madrid, E-28040, Spain.
Address Correspondence to: Dr. Ester Simonetti, Av. San Martín 4453 C1417DSE, Buenos Aires, Argentina, e-mail: simonetti@agro.uba.ar ; phone: +54 11 4524 8061.
Recibido / Received 13.IX.2011.
Aceptado / Accepted 6.XII.2011.
Abstract. In a previous work, deduced amino acid sequences from twenty wheat peroxidase genes were assigned to seven groups designated as TaPrx108 to TaPrx114. Some of these apoplastic peroxidases have previously shown to play different roles in the plant defense responses to infection by the cereal cyst nematode Heterodera avenae. In the present study, PCR marker analysis using Sears's aneuploid wheat lines cv. 'Chinese Spring' was used to locate four genes encoding peroxidase isozymes. The TaPrx111-A, TaPrx112-D and TaPrx113-F genes were located on the short arm of chromosome 2B and the TaPrx109-C on the long arm of chromosome 1B. These results would agree with the synteny between wheat and rice chromosomes previously established in other studies.
Keywords: Triticum aestivum; Synteny; Homologous; Chromosomal location.
Resumen. En un estudio previo se caracterizaron, alinearon y relacionaron 20 genes que codifican para peroxidasas en trigo. Las proteínas resultantes se asignaron a siete grupos, denominados TaPrx108 a TaPrx114, de acuerdo a la similitud entre ellas y con peroxidasas de Clase III de otras especies. En líneas de trigo resistentes al nemátodo Heterodera avenae, se ha demostrado que algunas de estas peroxidasas apoplásticas están relacionadas con las respuestas de defensa frente a la infección por dicho nematodo. En el presente estudio se localizaron cromosómicamente cuatro genes que codifican para enzimas peroxidasas mediante la reacción de PCR, utilizando pares de cebadores específicos para cada gen y ADN aislado de lineas de trigo aneuploides del cultivar Chinese Spring. Los genes TaPrx111-A, TaPrx112-D y TaPrx113-F fueron asignados al brazo corto del cromosoma 2B, mientras que el gen TaPrx109-C se localizó en el brazo largo del cromosoma 1B. Estos resultados mantendrían la sintenia previamente establecida entre los cromosomas de trigo y arroz.
Palabras clave: Triticum aestivum; Sintenia; Homólogos; Localización cromosómica.
INTRODUCTION
Plant peroxidases (EC 1.11.1.7), often designated as Class III peroxidases (Welinder, 1992) are heme containing proteins widely distributed in the plant kingdom. They are members of a large multigenic family, with 73 members in Arabidopsis (Tognolli et al., 2002) and 138 members in rice (Passardi et al., 2004). In contrast to several other gene families, peroxidase genes are twice as numerous in rice as in Arabidopsis. This large number of genes resulted from various duplication events by different mechanisms (Zhang, 2003): unequal crossing-over, various transposition events, duplication of large chromosome segments or polyploidization. This is reflected by the distribution of the peroxidase loci in clusters of closely homogeneous genes, among the 12 rice chromosomes (Passardi et al., 2004). Peroxidase genes arranged in tandem have also been found in many other plant species as horseradish (Fujiyama et al., 1988), wheat (Båga et al., 1995) and tomato (Roberts & Kolattukudy, 1989).
Peroxidase isozymes in hexaploid wheat have been extensively studied, and several homologous sets of loci controlling peroxidases have been identified independently by different researchers (Bosch et al., 1986, Liu et al., 1990).
In a previous work from our group, twenty peroxidase genes from wheat were identified and assigned to seven different groups (designated as TaPrx108 to TaPrx114). These represent peroxidases secreted to the apoplast by a putative N-terminal peptide signal. In addition, three groups of peroxidase genes (TaPrx111, TaPrx112 and TaPrx113) were found to be greatly induced by the cereal cyst nematode Heterodera avenae Woll. in the H-93-8 resistant wheat line (Simonetti et al., 2009).
MATERIALS AND METHODS
PCR marker analyses were used to determine the chromosomal location of peroxidase genes previously characterized by our group. Total DNA was isolated using the method described by Taylor & Powell (1982) from leaves of two-week old nulli-tetrasomic and ditelosomic series of Triticum aestivum L. cv. 'Chinese Spring' (Sears, 1954, 1966). Gene-specific primer pairs (generated from introns or flanking 5' or 3' untranslated regions) were designed using VECTOR NTI ADVANCE v.10 (Invitrogen) software and used to selectively amplify the different peroxidase genes (Table 1). PCR amplifications were performed in a Mastercycle Gradient thermal cycler (Eppendorf HQ, Germany). Amplification conditions were adjusted as recommended by the suppliers or according to the standard conditions described by Sambrook & Russell (2001), but varying the time and annealing temperature as a function of fragment size and primers characteristics. Reactions were carried out in a 0.02 cm3 reaction mix containing 1×PCR buffer; 2 mM MgCl2; 1 U AmpliTaq Gold DNA polymerase (Applied Biosystems); 200 µM of each dATP, dTTP, dCTP, dGTP; 0.25 µM of each primer and approximately 20 ng of template DNA. The amplification products were analyzed by electrophoresis in 1-2% agarose gel in 1× tris acetate ethylendiaminetetracetic acid buffer and visualized by UV fluorescence after staining with ethidium bromide.
Table 1. Primer pairs employed for chromosomal location of peroxidase genes in 'Chinese Spring' aneuploid wheat lines, size of PCR products and annealing temperature (AT) used.
Tabla 1. Pares de cebadores utilizados para la localización de genes de peroxidasas en líneas de trigo aneuploides del cultivar Chinese Spring, tamaño de los productos amplificados por PCR y temperaturas de hibridación (AT) empleadas.

RESULTS AND DISCUSSION
PCR amplification experiments indicated that TaPrx111- A, TaPrx112-D and TaPrx113-F peroxidase genes are located on the short arm of chromosome 2B. This location can be deduced from lack of bands for nulli-tetrasomic N2B-T2D and ditelosomic 2BL (Fig. 1A, 1B and 1C, respectively). These three genes belong to the peroxidase groups previously characterized as being inducible by nematode attack (Simonetti et al., 2009) and could be linked each other.
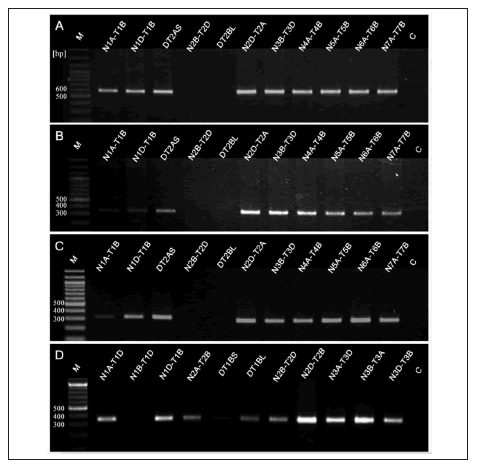
Fig. 1. Electrophoretic analysis of PCR products obtained with specific primers for TaPrx111-A (A); TaPrx112-D (B); TaPrx113-F (C) and
TaPrx109-C (D) genes, using genomic DNA from aneuploid wheat lines. M - 100 bp marker DNA ladder, C - control PCR without template.
Fig. 1. Análisis electroforético de productos de PCR obtenidos con cebadores específicos para los genes TaPrx111-A (A); TaPrx112-D (B);
TaPrx113-F (C) y TaPrx109-C (D), utilizando ADN genómico de líneas aneuploides de trigo. M - marcador de peso molecular con escalera de
100 pb, C - control de PCR sin ADN molde.
Based on sequence analysis, TaPrx111-A was determined to have high levels of identity with OsPrx112 gene in Oryza sativa, whereas TaPrx112-D and TaPrx113-F exhibited 77% identity to the rice gene OsPrx114 (Simonetti et al., 2009). These two rice peroxidases were earlier reported on rice chromosome 7 (Passardi et al., 2004), that is syntenous to wheat group 2 chromosomes (La Rota et al., 2004). These results would agree with the synteny established between chromosomes of wheat and rice (Gale & Devos, 1998).
Based on isoelectric point (pI) predictions, mature proteins represented either acid TaPrx111-A, TaPrx113-F or basic TaPrx112-D isoforms of peroxidases, with pIs ranging from 5.78, 5.79 to 8.82, respectively (Simonetti et al., 2009). In a previous work Liu et al. (1990) identified five sets of genes encoding peroxidase isozymes by isoelectric focusing (IEF) of extracts from different tissues of hexaploid wheat cv. 'Chinese Spring'. The Per-2 group, carried on the short arms of group 2 chromosomes, showed the highest activity in root tissues with stained isozymes focusing around pH 9.0. Nullisomic analysis of root extracts, demonstrated that two of the Per-2 isozymes (designated as '3' and '4'), were controlled by a gene(s) on the short arm of chromosome 2B. It is possible that one of these peroxidases would match with TaPrx112-D gene product (pI 8.82), given its overlapping expression pattern, pI and chromosomal location.
Electrophoretic analysis of PCR products obtained with specific primers for TaPrx109-C showed the lack of bands for nulli-tetrasomic N1B-T1D and ditelosomic 1BS (Fig. 1D), indicating that this peroxidase gene is located on the long arm of chromosome 1B. BLAST of TaPrx109 group sequences showed high identity to GenBank EST isolated from wheat plants under abiotic stress; however the biological function of this group is still unknown (Simonetti et al., 2009). All these genes showed high identity to rice peroxidase OsPrx74 at the DNA level although they lacked the third intron characteristic of this gene. The OsPrx74 locus has been identified on the rice chromosome 5 (Passardi et al., 2004). The location of TaPrx109-C gene on chromosome 1B would also agree with the synteny established between chromosomes of wheat and rice (Gale & Devos, 1998).
ACKNOWLEDGEMENTS
This study was supported by grants from the Comunidad Autónoma de Madrid (CAM-UPM Q09 0210-032). The authors are grateful to Mrs. M. López, C. Martínez-Belinchón and R. Rodriguez-Ríos for their technical assistance.
REFERENCES
1. Båga, M., R.N. Chibbar & K.K. Kartha (1995). Molecular cloning and expression analysis of peroxidase genes from wheat. Plant Molecular Biology 29: 647-662. [ Links ]
2. Bosch, A., A.M. Figueiras, M.T Gonzales-Jeans & C. Benito (1986). Leaf peroxidase-A biochemical marker for the group 2 chromosomes in the Triticinae. Genetical Research (Camb.) 47: 103-107. [ Links ]
3. Fujiyama, K., H. Takemura, S. Shibayama, K. Kobayashi, J.K. Choi, A. Shinmyo, M. Takano, Y. Yamada & H. Okada (1988). Structure of the horseradish peroxidase isozyme C genes. European Journal of Biochemistry 173: 681-687. [ Links ]
4. Gale, M.D. & K.M. Devos (1988). Plant comparative genetics after 10 years. Science 282: 656-659. [ Links ]
5. La Rota, M. & M.E. Sorrells (2004). Comparative DNA sequence analysis of mapped wheat ESTs reveals the complexity of genome relationships between rice and wheat. Functional and Integrative Genomics 4: 34-46. [ Links ]
6. Liu, C.S., S. Chao & M.D. Gale (1990). The genetical control of tissue-specific peroxidase, Per-1, Per-2, Per-3, Per-4 and Per-5 in wheat. Theoretical and Applied Genetics 79: 305-315. [ Links ]
7. Passardi, F., D. Longet, C. Penel & C. Dunand (2004). The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry 65: 1879-1893. [ Links ]
8. Roberts, E. & P.E Kolattukudy (1989). Molecular cloning, nucleotide sequence, and abscisic acid induction of a suberization-associated highly anionic peroxidase. Molecular and General Genetics 217: 223-232. [ Links ]
9. Sambrook, J. & D.W. Russell (2001). Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, 3rd ed., New York. [ Links ]
10. Sears, E.R. (1954). The aneuploids of common wheat. Missouri Agricultural Experiment Station Publications 572: 1-58. [ Links ]
11. Sears, E.R. (1966). Chromosome mapping with the aid of telocentrics. In: Mackey, J. (ed.): Proc. 2nd Int. Wheat Genet. Symp. Hereditas 2: 370-381. [ Links ]
12. Simonetti, E., P. Veronico, M.T. Melillo, A. Delibes, M.F. Andrés & I. López-Braña (2009). Class III Peroxidase characterization and expression pattern in a cereal cyst nematode (Heterodera avenae) resistant hexaploid wheat line. Molecular Plant-Microbe Interactions 22: 1081-1092. [ Links ]
13. Taylor, B. & A. Powell (1982). Isolation of plant DNA and RNA. Focus 4: 4-6. [ Links ]
14. Tognolli, M., C. Penel, H. Greppin & P. Simon (2002). Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 288: 129-138. [ Links ]
15. Welinder, K.G. (1992). Plant peroxidases: structure-function relationships. In: Penel, C., Gaspar, T. and Greppin, H. (eds), pp. 1-24. Plant Peroxidases. University of Geneva, Switzerland. [ Links ]
16. Zhang, J. (2003). Evolution by gene duplication: an update. Trends in Ecology and Evolution 18: 292-298. [ Links ]














