Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
Kurtziana
versión On-line ISSN 1852-5962
Kurtziana vol.37 no.1 Córdoba ene./jun. 2012
ARTÍCULOS ORIGINALES
A new species of Laetiporus (Basidiomycota, Polyporales) from the Caribbean basin
Mark T. Banik 1, 3, Daniel L. Lindner 1, Beatriz Ortiz-Santana 1 & Deborah J. Lodge 2
1 US Forest Service, Northern Research Station, Center for Forest Mycology Research, One Gifford Pinchot Drive, Madison, Wisconsin 53726, USA.
2 US Forest Service, Northern Research Station, Center for Forest Mycology Research, International Institute for Tropical Forestry, Luquillo, PO Box 1377, Puerto Rico 00773, USA.
3 Corresponding author: mbanik@fs.fed.us
Summary
Laetiporus caribensis is described as a new species, causing brown rot on hardwoods in the Caribbean basin. Laetiporus caribensis is distinguished from described species in the genus by smaller basidiospores. In addition, single spore isolates of L. caribensis are incompatible with those of the known species of Laetiporus from North America and Japan. The ITS sequence of L .caribensis is divergent from other members of the genus, being at least 50 base pairs different from L. gilbertsonii, the most closely related species.
Key words: Basidiomycota; Fungal phylogenetics; Fungi; Polyporaceae; Sulfur shelf.
Resumen
Una especie nueva de Laetiporus (Basidiomycota, Polyporales) de la cuenca del Caribe
Laetiporus caribensis se describe como una nueva especie dentro de este género, causando una pudrición castaña en angiospermas en la cuenca del Caribe. Laetiporus caribensis se distingue de la mayoría de las otras especies en el género por tener basidiosporas más pequeñas. Además, los cultivos monospóricos de L. caribensis son incompatibles con los de las especies conocidas de Laetiporus de América del Norte y Japón. La secuencia de ITS de L. caribensis es divergente de las de otros miembros del género, teniendo por lo menos 50 bp diferentes de la de L. gilbertsonii, la especie más estrechamente relacionada.
Palabras clave: Basidiomycota; Filogenética de hongos; Fungi; Polyporaceae.
Introduction
Recent reports have described several new taxa previously contained within the Laetiporus sulphureus (Bull.:Fr.) Murill species complex (Banik et al. 1998, Burdsall & Banik 2001, Ota et al. 2009, Tomsovský & Jankovský 2008). In North America, five species distinguished by single spore isolate (SSI) incompatibility reactions in culture, ITS sequences and morphological and ecological characteristics are now recognized. The ITS sequences of these species each fall in unique clades corresponding to their species designation (Lindner and Banik 2008). Likewise, two Asian species and one Asian/European species also possess ITS sequences that belong to unique clades and also form unique incompatibility groups in culture (Ota & Hattori 2008, Ota et al. 2009, Banik et al. 2010). Based on these reports, the link between incompatibility group designation and clade level ITS phylogeny is well established, allowing ITS-based data to be used as a reliable indicator of species groups.
Several reports define ITS sequence clades that do not coincide with any described species in Laetiporus (Lindner and Banik 2008, Vasaitis et al. 2009, Banik et al. 2010). One of these, designated Clade J by Vasaitis et al. (2009), originates from collections obtained from several islands in the Caribbean basin. Based on ITS sequence, Clade J is the most distinctive of all the western hemisphere clades thus far elucidated. However, it still possesses the classic Laetiporus characteristics of orangish-yellow, laterally stipitate shelved fruiting bodies, lacking clamp connections and causing a brown rot. In addition both ITS and LSU sequences place it in the core Laetiporus clade (Lindner and Banik 2008). The purpose of this study is to define the unique characteristics of Clade J and to describe this taxon as a new species within Laetiporus.
Materials and Methods
Specimens and microscopic examinations-Ten collections were examined: four from Puerto Rico, three from Costa Rica, two from Argentina and one from Belize. The collection information for these specimens is provided in the remarks section of the new species description. Microscopic features were studied by placing thin, freehand sections in 3% KOH and/or stained with 1.0% phloxine and observed in oil immersion under a compound microscope.
DNA sequencing and phylogenetic analysis- Nuclear ITS rDNA from fruiting bodies was amplified from hymenial tissue using the technique of Lindner and Banik (2009). The technique was modified by replacing the EXO/ SAP cleanup of the PCR products with an approximately 1:10 dilution of the products prior to use in sequencing. Sequencing of amplicons and maximum parsimony analyses followed the protocols of Lindner and Banik (2008), while the maximum likelihood analyses followed Lindner and Banik (2011). Sequences not matching previously identified Laetiporus species were aligned with a reference database of previously identified Laetiporus ITS sequences (Banik et al. 2010) using ClustalX 2.1 (Larkin et al. 2007). New sequences were deposited in GenBank (Benson et al. 2011).
Single spore isolate pairings-Single spore isolates were obtained from one fruit body, PR-6521, following the technique of Banik and Burdsall (1999), except that the basidiospores were germinated on potato dextrose agar (Difco) with pH lowered to approximately 3.5 with the addition of 1M HCl. To determine the within fruiting body incompatibility system, twenty sibling SSI from PR-6521 were paired against each other in all combinations. The pairings were conducted and evaluated on the basis of morphological changes according to the technique described by Banik and Burdsall (1999). To determine the relationship of PR-6521 to other established Laetiporus species, four SSIs of PR-6521 were paired against tester SSIs of four North American and three Japanese species.
Results
Sequence analysis-The ITS regions of 10 fruiting bodies were amplified and sequenced. Five collections, four from Puerto Rico (PR- 914, PR-6317, PR-6521, PR-6583) and one from Costa Rica (Welden 3174), possessed the 555 bp sequence of Clade J. Another specimen from Costa Rica (Muez 207) had eight single bp changes when compared to Clade J, and a length of 563 bp due to a single insertion of nine bp and an additional deletion of a single base pair. Based on our interpretation of Laetiporus phylogeny, this specimen represents a new ITS clade, designated Clade L. Another new ITS clade, Clade M, is represented by Robledo 1122 from Argentina and has an ITS length of 572 bp. Compared to Clade J it has 11 single bp changes, two separate nine bp insertions, a one and a two bp insertion, and a one and a three bp deletion. Clade L and M differ by 10 bp changes, a nine bp insertion/deletion, and two separate three bp insertions/deletions. Three collections, one each from Argentina (Robledo 47), Belize (BZ-544), and Costa Rica (Lowe-12954), were identified as L. gilbertsonii Burds. based on 100% ITS sequence match (GenBank accession numbers JN684769, JN684768, JN684767, respectively). Because L. gilbertsonii is well characterized both morphologically and phylogenetically, these specimens and their associated ITS sequences were not considered further.
The maximum parsimony and maximum likelihood analyses produced similar overall results; the addition of Clades L and M expanded the number of known Laetiporus clades to 14 (Figure 1). The uniqueness of Clade J was not compromised by the addition of the new clades L and M, and the uniqueness of Clade J from the other identified clades was further strengthened by the presence of clades L and M.
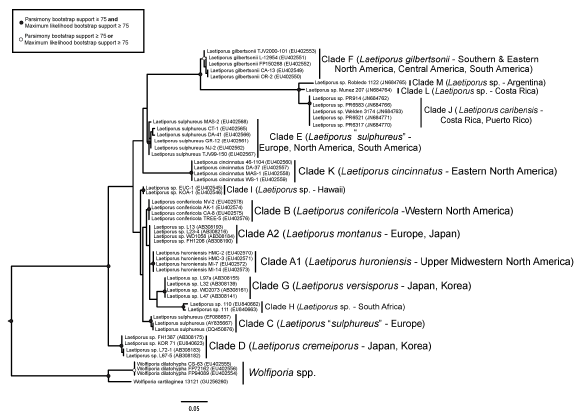
Fig. 1. Maximum likelihood phylogeny of all known Laetiporus clades based on ITS rDNA sequences. Species clades are indicated with vertical black lines and clade names follow Banik et al (2010).
Pairing tests-Forty three percent of the 190 PR-6521 sibling pairings were rated as positive, 49 % fused and were rated negative, and in the remainder of the pairings the two test SSIs remained separate without the formation of a line. The pattern of the reactions observed did not fit into a readily recognizable mating system.
All 80 pairings between four SSIs of PR- 6521 and four SSIs tester isolates each of L. cincinnatus (Morgan) Burds., Banik & Volk, L. sulphureus clade E, L. gilbertsonii, L. montanus Cerny ex Tomsovsky & Jankovsky and L. versisporus (Lloyd) Imazeki were characterized by a separation of the test isolates by a dark line (DL) indicative of a negative reaction. Negative reactions were also observed in the 20 pairings between five SSIs of L. cremieporus Y. Ota & T. Hatt. and the PR-6521 SSIs . The reactions observed between the four L. conifericola Burds. & Banik SSI and those of PR-6521 were less definitive. Five of the reactions exhibited DL formation and thus were rated negative. The remainder exhibited a reaction type not previously documented. It was characterized by the formation of a barrage zone between the two test isolates but no DL was formed. Since the reaction zone did not express an increase in density and pigmentation associated with a positive reaction, these reactions were also rated as negative.
Taxonomy
Laetiporus caribensis Banik & D. L. Lindner, sp. nov. Figs. 2-3.

Fig. 2. Fruiting body of Laetiporus caribensis (holotype, PR-6583).
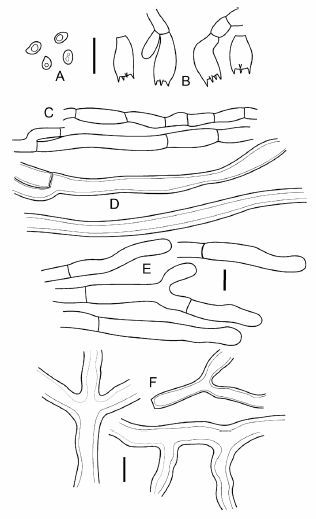
Fig. 3. Microscopic features of Laetiporus caribensis (holotype, PR-6583). A. basidiospores, with oil droplets; B. basidia; C. generative hyphae of pore trama; D. skeletal hyphae of pore trama; E. hypha of pileus surface; F. hypha of pileus context. Scale bar = 10 µm.
Mycobank: MB 563162
Species haec ut Laetiporus sulphureus (Bull.:Fr.) Murrill, sed basidiosporis eliposoidiis minoris 4.0-4.5 x 2.7-3.6 µm (ovoidiis 5.5-7 x 4-5 µm in L. sulphureus).
Holotype: PUERTO RICO. El Yunque National Forest, Luquillo Mountains, D. Jean Lodge, PR-6583, 17-XI-2009."on Guarea guidonia snag" Holotypus speciei (CFMR).
Topotype: D. Jean Lodge PR-6521, 6-III- 2008 (CFMR).
Isotype: D. Jean Lodge PR-6583, 17-XI- 2009 (UPR-RP).
Basidiomata annual, pileus dimideate, imbricate clusters, 15-20 cm broad, upper surface zonate orange/pale orange transitioning to pale orange/yellow when young zonation fading when dry, mature surface dull, tuberculate; context soft, easily broken, white, pale orangish buff tint near attachment; pore surface lemon yellow, pores minute, subangular, 4-5/mm, 1 mm deep. Odor not distinctive, taste none.
Pileus surface hyphae erect 5.4-11 µm diameter, hyaline, thin to moderately thick-walled, smooth. Pore trama dimitic, composed of generative and skeletal hyphae. Generative hyphae parallel, 2.7-5.4 µm diameter, hyaline, frequently septate, lacking clamp connections. Skeletal hyphae subparallel, 3.6-5.9 µm diameter, hyaline, rarely septate, lacking clamp connections, walls 0.5-1.4 µm thick. Context binding hyphae 6.3-12 µm diameter, walls 1.4- 2.7 µm thick.
Hymenium of basidia. Basidia clavate, 8.6-15.3 x 5.4-6.3 µm, hyaline, thin walled, 4 sterigmate, lacking a basal clamp. Basidiospores ellipsoid, 4-4.5 x 2.7-3.6 µm, hyaline, thin walled, smooth, guttulate.
Substrate: causing a brown heart rot of tropical hardwoods.
Distribution: on islands in the Caribbean basin and in Central America.
Etymology: known from the Caribbean basin.
Material examined: COSTA RICA. Alajuela Province, A. L. Welden 3174, 30-IX-1972. "on decorticated log" (CFMR). PUERTO RICO. Luquillo: Caribbean National Forest, El Verde Research Area, S. Cantrell and P. Melendez PR-914, 21-VII- 1992."on hardwood tree stump" (CFMR). PUERTO RICO. Luquillo: Caribbean National Forest, El Verde Research Area, M. Santana PR- 6317, 15-XI-2000"on Dacroydes excelsea" (CFMR).
Additional material examined: For comparative purposes the following specimens were examined: L. gilbertsonii-ARGENTINA. Jujuy Province, El Fuerte, Sierra de Santa Bárbara, Gerardo Robledo 47, 23-II- 2001."on live stem" (CORD). BELIZE. Cayo District, Mountain Pine Ridge Forest Preserve, Leif Ryvarden BZ-544, 19-IX-2001"on log" (BRH). COSTA RICA. San Jose Province, El Empalme, J.L. Lowe 12954, 28-VI-1963."on hardwood" (CFMR). Laetiporus Clade L-COSTA RICA. Guanacaste Province, Parque Nacional Guanacaste, Area de Conservacion Guanacaste, M. Muez 207, 5-VIII-1993 (CFMR). Laetiporus Clade M-ARGENTINA. Salta Province, Parque Nacional El Rey, Gerardo Robledo 1122, 26- III-2007."on live tree trunk" (CORD).
Discussion
Laetiporus caribensis is similar in color and other macroscopic characteristics to most other species in the genus. Microscopically it is distinct in having spores approximately two-thirds the size of L. cincinnatus, which possesses the next smallest spores in the genus. Other microscopic characters such as the presence of binding hyphae and basidia size are congruent with other members of the genus. The ITS sequence of L. caribensis is distinct from all the other known species. Furthermore, the pairing data demonstrate L. caribensis is distinct from the other species of the genus for which SSIs are available. However, the nature of the incompatibility system within the species could not be determined from the single collection of SSIs available for study.
Laetiporus caribensis joins L. gilbertsonii as the only confirmed Laetiporus species present on islands in the Caribbean. The range of L. gilbertsonii also extends into the southern and western parts of the United States and northwestern Argentina. These two species have similar fruiting habits but are easily separated based on differences in spore size as well as distinctive ITS sequences. Laetiporus cincinnatus is not known outside of continental North America, but has been found as far south as Louisiana; therefore it is possible that its range could overlap with that of L. caribensis. However, L. cincinnatus has a white pore layer and forms a rosette while fruiting from the soil and thus is easily separated from L. caribensis. No other described species of Laetiporus has a geographical distribution similar to that of L. caribensis.
The status of L. caribensis in relation to Clades L and M needs to be more fully examined. Each of these clades is represented by one specimen and thus assessing their geographical range is not possible. Based on ITS sequence analysis, both Clade L and M are closely related to L. caribensis and they all have basidiospores of similar size. Clades J (L. caribensis), L and M are more closely related to each other than the next closest clade, Clade F (L. gilbertsonii). It is possible Clades L and M and L. caribensis are variants of the same species, but based on the current understanding of ITS phylogeny in the genus, their sequences are dissimilar enough to make this seem unlikely. More collections of members of these clades, especially from regions of Central and South America, are needed to fully assess their relationship to each other and L. caribensis. Collections of SSIs of all these taxa would also be useful in determining their taxonomic status and incompatibility systems.
Acknowledgements
We would like to acknowledge Karen Nakasone for help in Latin name selection and Gerardo Robledo for providing specimens from Argentina.
References
1. Banik M. T. & H. H. Burdsall Jr. 1999. Incompatibility between Laetiporus cincinnatus and L. sulphureus in culture. Mycotaxon 70:461-469. [ Links ]
2. Banik M. T.,H. H. Burdsall Jr., & T. J. Volk. 1998. Identification of groups within Laetiporus sulphureus in the United States based on RFLP analysis of the nuclear ribosomal DNA. Folia Cryptog Estonica 33:9-14. [ Links ]
3. Banik M. T., D. L. Lindner, Y. Ota & T. Hattori. 2010. Relationships among North American and Japanese Laetiporus isolates inferred from molecular phylogenetics and single-spore incompatibility reactions. Mycologia 102:911-917. [ Links ]
4. Benson D.A., I. Karsch-Mizrachi, D.J. Lipman, J. Ostell,& E.W. Sayers. 2011. GenBank. Nucleic Acids Research 39:D32-7. [ Links ]
5. Burdsall H. H. Jr., & M. T. Banik. 2001. The genus Laetiporus in North America. Harvard Papers in Botany 6:43-55. [ Links ]
6. Larkin M.A., G. Blackshields, N.P. Brown, R. Chenna, P.A. McGettigan, H. McWilliam, F. Valentin, I.M. Wallace, A. Wilm, R. Lopez, J.D. Thompson, T.J. Gibson, D.G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947-2948. [ Links ]
7. Lindner D. L. & M. T. Banik. 2008. Molecular phylogeny of Laetiporus and other brown-rot genera in North America. Mycologia 100:417-430. [ Links ]
8. Lindner D. L. & M. T. Banik. 2009. Effects of cloning and root-tip size on observations of fungal ITS sequences from Picea glauca roots. Mycologia 101:157-165. [ Links ]
9. Lindner D. L. & M. T. Banik. 2011. Intragenomic variation in the ITS rDNA region obscures phylogenetic relationships and inflates estimates of operational taxonomic units in genus Laetiporus. Mycologia 103:731-740. [ Links ]
10. Ota Y., & T. Hattori. 2008. Relationships among three Japanese Laetiporus taxa based on phylogenetic analysis and incompatibility tests. Mycoscience 49:168-177. [ Links ]
11. Ota Y., T. Hattori, M. T. Banik, G. Hagedorn, K.Sotome, S. Tokuda & Y. Abe. 2009. The genus Laetiporus (Basidiomycota, Polyporales) in east Asia. Mycol. Res 113:1283-1300. [ Links ]
12. Tomsovský, M. & L. Jankovský. 2008. Validation and typification of Laetiporus montanus. Mycotaxon 106:289-295. [ Links ]
13. Vasaitis R., A. Menkis, Y. W. Lim, S. Seok, M. Tomsovský, L. Jankovský, V. Lygis, B. Slippers & J. Stenlid. 2009. Genetic variation and relationships in Laetiporus sulphureus s. lat., as determined by ITS rDNA sequence and in vitro growth rate. Mycol. Res 113:326-336. [ Links ]
Original recibido el 8 de Julio de 2011;
primera decisión: 18 de Agosto de 2011;
aceptado el 22 de Septiembre de 2011.
Editor responsable: Gerardo Robledo.














