Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
Kurtziana
versión On-line ISSN 1852-5962
Kurtziana vol.37 no.1 Córdoba ene./jun. 2012
ARTÍCULOS ORIGINALES
Rickiella edulis and its phylogenetic relationships within Sarcoscyphaceae
Andrea I. Romero 1, Gerardo Robledo 2, Katherine F. LoBuglio 3, 4 & Donald H. Pfister 4
1 PROPLAME-PRHIDEB-CONICET. Departamento de Biodiversidad y Biología Experimental. Facultad de Ciencias Exactas y Naturales, Universidad de Buenos Aires. Piso 4°, Pabellón II, Lab 5, Int. Güiraldes 2620. Ciudad Universitaria. C1428EHA, Buenos Aires, Argentina. E-mail: romero@bg.fcen.uba.ar
2 Laboratorio de Micología, IMBIV Instituto Multidisciplinario de Biología Vegetal, CONICET, Universidad Nacional de Córdoba. C.C. 495, 5000, Córdoba, Argentina. E-mail: glrobledo@yahoo.com
3 E-mail: klobuglio@oeb.harvard.edu
4 Farlow Herbarium, Harvard University Herbaria, 22 Divinity Ave., Cambridge, Massachusetts 02138, USA. E-mail: dpfister@oeb.harvard.edu
Summary
Rickiella edulis is reported from Argentina for the first time and is documented with photographs of fresh specimens and molecular data. Previously the species was known as R. transiens (= Phillipsia transiens) and was reported from southern Brazil and Paraguay. Phylogenetic analyses based on SSU rDNA and LSU rDNA shows its placement in a monophyletic family, the Sarcoscyphaceae. The relationship of Rickiella, Phillipsia and Nanoscypha however could not be resolved from phylogenetic analyses of the ITS, SSU, and LSU rDNA sequences. The excipular tissue of Rickiella is shot through with regularly spaced channels and cavities. Because of this feature, the genus Rickiella is recognized as distinct from Phillipsia. Phillipsia and Nanoscypha are morphologically distinct but diversity within Phillipsia remains a topic for further research. A new tribe in the Sarcoscyphaceae is proposed to accommodate the genus Wynnea.
Key words: Argentina; Nanoscypha; Pezizales; Phillipsia; Phylogeny.
Resumen
Rickiella edulis y sus relaciones filogenéticas dentro de las Sarcoscyphaceae
Rickiella edulis se registra por primera vez para la Argentina y se documenta a través de fotografías de materiales frescos y de datos moleculares. Originalmente la especie fue conocida como R. transiens (= Phillipsia transiens) y registrada para Brasil y Paraguay. Análisis filogenéticos basados en los marcadores SSU y LSU muestran su ubicación en la familia monofilética, Sarcoscyphaceae. Sin embargo, las relaciones entre los géneros Rickiella, Phillipsia y Nanoscypha no se pudieron resolver a partir del análisis filogenético basados en los marcadores de ITS, SSU y LSU rDNA. Las características particulares del excípulo de Rickiella, lacunoso y con cavidades, lo diferencian de Phillipsia. Phillipsia y Nanoscypha son morfológicamente distinguibles pero la diversidad dentro de Phillipsia es un tema para futuras investigaciones. Se propone una nueva tribu dentro de Sarcoscyphaceae para acomodar el género Wynnea.
Palabras clave: Argentina; Nanoscypha; Pezizales; Phillipsia; Filogenia.
Introduction
A recent collection of Rickiella edulis (Speg.) Pfister from NW Argentina renewed interest in the status of this unusual taxon and in the known distribution of the species. Pfister (1987), in publishing on his discovery of the older name for R. transiens Sydow, reviewed the small literature then available regarding the genus and the single species recognized in it. To this review there is nothing new to add. Korf (1983) in an earlier study considered Rickiella to be a synonym of Phillipsia from which it differs in possessing an excipulum with many regularly spaced lacunae or cavities. None of the phylogenetic studies involving the family Sarcoscyphaceae and related families in the Pezizales have included this species (Hansen and Pfister 2006, Harrington et al. 1999, Perry et al. 2007, Pfister et al. 2008). This recent collection allows us to review the morphology, to consider the distribution of the species, and to sample DNA for phylogenetic study.
The history of the family Sarcoscyphaceae has been reviewed by Harrington et al. (1999). No suitable material of R. edulis was available at that time. That study was based on SSU rDNA (Small Subunit Ribosomal DNA) sequences and had limited samples, generally with only a single example for each genus. Harrington et al. (1999) and subsequent studies have demonstrated a monophyletic family Sarcoscyphaceae. Based on Harrington's study, Nanoscypha was used as an outgroup species by Hansen et al. (1999) in their studies of Phillipsia using ITS rDNA (Internal Transcribed Spacer) sequences.
Using ITS, SSU, and LSU rDNA (Large Subunit Ribosomal DNA) we have returned to investigate the relationships among members of the Sarcoscyphaceae in an attempt to elucidate the placement of Rickiella particularly with reference to Phillipsia, the genus in which Korf (1983) suggested the type species of Rickiella might be placed. We also are able to elaborate on the distribution of R. edulis.
Materials and Methods
Herbarium Specimens Used for Morphological Evaluation and DNA Samples
Specimen for DNA extraction: Rickiella edulis, collected by Gerardo Robledo in 2007, was sent to D. Pfister by A. I. Romero from BAFC Mycotheca (BAFC #51697). Specimens examined: ARGENTINA, Prov. Salta, La Caldera, Camino de tierra que une Ruta 9 (Camino de Cornisa) con General Güemes, 24º40'20.5''S, 65º22'4.8''W, on dead fallen logs, 1349 m asl, Robledo 871, 872, 873, 20-II-2007 (CORD) and #51698/99 (BAFC) (acronyms according to Thiers, 2011).
Kompsoscypha phyllogena (Seaver) Pfister was collected by D.J. Lodge and L. Millman in 2009 (without other data) at El Yunque, Baisley Watershed, Puerto Rico (FH #DHP 10-690). Two specimens of Phillipsia (FH #113 and #114) were collected in the Dominican Republic (Jardín Botanico Nacional"Dr. Rafael M. Moscoso" Santo Domingo, Republica Dominicana, without data) by S. Cantrell et al. (2002). Specimen #114 was confirmed to be P. crispata based primarily on spore morphology and 95% ITS rDNA sequence similarity to P. crispata (GenBankAF117355, T. Læssøe AAU-44895a, and GenBank AF117354, T. Læssøe AAU-44801, Hansen et al. 1999). Specimen #113 was confirmed to be P. carnicolor based on 97% ITS rDNA sequence similarity to P. carnicolor (GenBank AF117353, D. Pfister DHP-7126, Hansen et al. 1999).
DNA Samples: DNA samples of Rickiella were obtained from the specimen of R. edulis (Robledo 873/BAFC 51697, see above) sent by A.I. Romero. DNA samples of the Phillipsia species used in this expanded study were obtained from K. Hansen (Naturhistoriska Riksmuseet Stockhom, Sweden) and were the same genomic DNA samples used in a previous study by Hansen et al. (1999): Phillipsia carnicolor D. Pfister DHP-7126, P. olivacea Halling-5456, P. olivacea T. Læssøe AAU-43162, P. lutea G.J. Samuels and P. Searwar NY-4113, and P. domingensis D. Pfister DHP-7169. These DNA samples were used in amplification of the LSU and SSU rDNA region. A DNA sample of Pithya cupressina, which originated from a previous study by Harrington et al. (1999), was used to amplify the LSU rDNA region.
DNA Isolation, PCR, and Sequencing Techniques
DNA was extracted from the herbarium specimen of R. edulis (#51697) and K. phyllogena (DHP 10-690) using the Qiagen DNeasy Plant Mini Kit (cat. no. 69104). A 1/10 and 1/100 dilution of the DNA was used for PCR amplification of the ITS, SSU and LSU rDNA regions. The ITS rDNA region was amplified using ITS1F (Gardes and Bruns 1993) and ITS4 (White et al. 1990). PCR parameters were as previously described (LoBuglio et al. 1993), using 35 PCR amplification cycles. The SSU was amplified using the NS1, NS2, NS4, NS8 (White et al. 1990) and SL1, SL122, SL344 (Landvik et al. 1996) primers. Amplification of the LSU rDNA region utilized the primers LROR and LR5 (Monclavo et al. 2000). All PCR reactions were done in a Peltier Thermal cycler PTC-200 (MJ Research, Watertown, MA), and used EconoTaq DNA Polymerase (Lucigen, Middleton, WI). PCR amplification, purification, and sequencing techniques were as described in Hansen et al. (2005). Sequencher 4.6 (GeneCodes, Ann Arbor, Michigan) was used to edit the DNA sequences obtained. The DNA sequences determined in this study were deposited in GenBank (R. edulis ITS=JQ260808, LSU=JQ260809, SSU=JQ260819; K. phyllogena LSU=JQ260810, SSU=JQ260820; P. carnicolor DHP- 7126 LSU=JQ260811, SSU=JQ260821; P. carnicolor #113 LSU=JQ260812, SSU=JQ260822; P. crispata #114 LSU=JQ260813, SSU=JQ260823; P. olivacea Halling-5456 LSU=JQ260814, SSU=JQ260824; P. olivacea AAU-43162 LSU=JQ260815, SSU=JQ260825; P. lutea NY-4113 LSU=JQ260816, SSU=JQ260826; P. domingensis DHP-7169 LSU=JQ260817, SSU=JQ260827; and Pithya cupressina LSU=JQ260818).
DNA Sequence Analyses
Alignment of the DNA sequences was done using Se-Al v 2.0a8 (Rambaut 1996).
As previously described (LoBuglio and Pfister 2010), DNA sequence alignments were analyzed using: MrBayes v3.0b4 (Ronquist and Heulsenbeck 2003) for obtaining Bayesian posterior probabilities (PP); Maximum Parsimony using PAUP 4.0b10 (MP; Swofford 2002); and Maximum-Likelihood with RAxML-HPC2 on Abe through the Cipres Science Gateway (ML; Miller et al. 2009). Branch support for MP and ML analyses was determined by 1000 bootstrap replicates.
The ITS sequence of Rickiella obtained in this study was aligned with ITS sequences of the Phillipsia and Nanoscypha species included in the study by Hansen et al. (1999) (study S403 TreeBASE, http://purl.org/phylo/treebase/phylows/study/TB2:S403). Sarcoscypha coccinea (DQ491486) and Sarcoscypha austriaca (U66012) were included in this analysis as outgroup species. The hypervariable ITS1 region (as described by Hansen et al. 1999) was aligned with multiple gaps. Parsimony analyses were carried out with these gapped positions either included or excluded, and followed a phylogenetic search protocol using Maximum Parsimony as outlined by Hansen et al. (1999).
The DNA sequences determined in this study (Rickiella, Kompsoscypha, Phillipsia species, and Pithya), were included in the SSU and LSU phylogenetic analysis along with the following sequences from GenBank (SSU and LSU respectively): Nanoscypha tetraspora AF006314+DQ220374, Pseudopithyella minuscula AF006317+AY544658, Sarcoscypha coccinea AY544691+AY544647, Microstoma floccosum AF006313+DQ220370, Cookeina tricholoma AF006311+AY945860, Pithya cupressina AF006316, Chorioactis geaster AF104340+AY307944, Wynnea sp./americana AF006319+AY945848, Wolfina aurantiopsis AF104664+AY945859, Desmazierella acicola AF104341+AY945854, Neournula pouchetii AF104666+AY307940, Sarcosoma latahense FJ176806+FJ176860, Urnula craterium AF104347.1+AY945851, and Galiella rufa AF004948+AY945850. The outgroup taxa were: G. rufa, and U. craterium.
A third data set, which combined the ITS, LSU, and SSU rDNA data, was constructed and analyzed. This data set included 12 taxa: P. domingensis DHP- 7169, P. lutea NY-4113, P. olivacea Halling-5456, P. olivacea AAU-43162, P. crispata #114, P. crispata AAU-44895a, P. carnicolor #113, P. carnicolor DHP-7126, Nanoscypha tetraspora, R. edulis, and Pithya cupressina and Sarcoscypha coccinea as the two outgroup species.
Hypothesis Testing
Approximately Unbiased (AU) tests (Shimodaira 2002; Ruhfel et al. 2008; Mathews et al. 2010) were conducted with the SSU and LSU rDNA data set using the R (http://www.r-project.org/) package, Scaleboot, to statistically evaluate alternative phylogenetic hypotheses on the evolution of Rickiella, Phillipsia and Nanoscypha. The three hypothesis tree topologies tested were: 1) All species of Phillipsia are monophyletic; 2) Nanoscypha and Phillipsia species are monophyletic; and 3) Rickiella and Phillipsia species are monophyletic. Constraint trees were first drawn in MacClade 4.05 (Maddison and Maddison 2004) to enforce the above mentioned tree topologies and then tested against the best ML tree.
Results
The ITS1F-ITS4 sequence of Rickiella was 613 bp long. It is alignable with the ITS of Phillipsia species from Hansen et al. (1999), but shows a region of 80 unique base pairs (bp) at approximately 69 bp in ITS1. In the study by Hansen et al. (1999), a hypervariable region among the ITS sequences of Phillipsia and Nanoscypha species at this same ITS1 region was described. Parsimony analysis of Rickiella, Phillipsia, and Nanoscypha ITS sequences (with S. coccinea and S. austriaca as the outgroup) showed that both Rickiella and Nanoscypha were unresolved within a highly supported (100%) Phillipsia clade (results not shown). Trees were identical whether gapped positions (present in the ITS1 hypervariable region) were included or excluded from the analyses as was previously found (Hansen et al. 1999).
The combined SSU and LSU alignment of the 24 taxa in Figure 1 included 2662 bp, none of which were excluded in the phylogenetic analyses. MP and ML bootstrap support and PP values for the family Sarcoscyphaceae was 100%. This was the case when the data set was analyzed 3 times, each using 3 different outgroups: 1) Urnula craterium and Galiella rufa; 2) Neournula pouchetii, Desmazierella acicola, Sarcosoma latahense, Chorioactis geaster, and Wolfina aurantiopsis; and 3) no outgroup. Tree topologies from Bayesian and ML analyses did not conflict with the parsimony tree presented in Figure 1, with respect to relationships within the Sarcoscyphaceae among statistically supported branches (= 60% MP, 70% ML, and 90% PP). As shown in Figure 1, Bayesian, MP, and ML phylogenies supported a monophyletic clade comprised of Rickiella, Nanoscypha and the five species of Phillipsia examined (MP had low support, 66%, compared to ML bootstrap, 90%, and Bayesian PP values, 100%). Within this clade the five species of Phillipsia, Rickiella, and Nanoscypha collapses to a polytomy. Inclusion of the ITS data, in the LSU, and SSU data set (as described in materials and methods) did not improve the phylogenetic resolution among the Phillipsia, Rickiella, and Nanoscypha species (data not shown). As described above for the LSU-SSU phylogeny presented in Figure 1, the ITS, LSU, and SSU data also supported a monophyletic clade (with 100% and 97% support, MP and ML Bootstrap respectively) comprised of Rickiella, Nanoscypha and the five species of Phillipsia examined. Within this clade there was no bootstrap support among the species of Phillipsia, Rickiella, and Nanoscypha. Thus, the relationship among the three genera, Rickiella, Phillipsia, and Nanoscypha, was not resolved from phylogenetic analyses of rDNA sequence data.
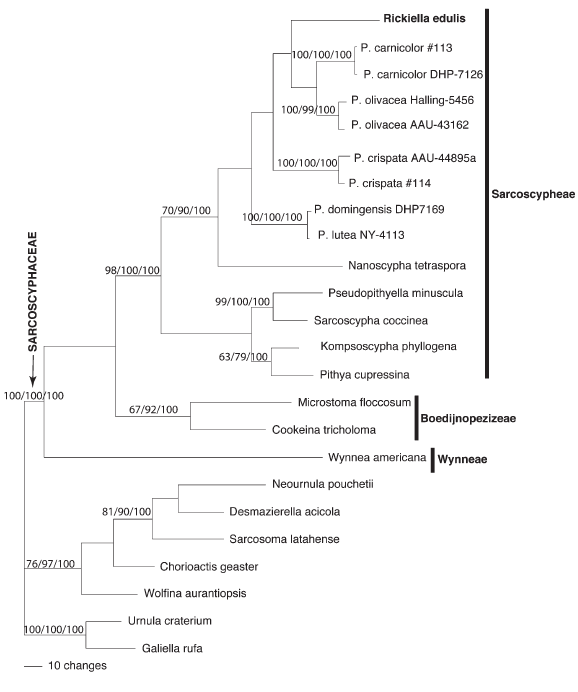
Fig. 1. One of 3 most parsimonious trees (tree length of 1015) based on LSU and SSU rDNA sequence data (2662 bp). Parsimony and Maximum-Likelihood Bootstrap values below 60%, and Bayesian posterior probabilities below 90% are not displayed. The outgroup taxa were, G. rufa, and U. craterium. The vertical line indicates tribes in the Sarcoscyphaceae: Sarcoscypheae, Boedijnopezizeae and Wynneae.
The AU test (Table 1) rejected hypothesis tree #1, which forced all species of Phillipsia to be a monophyletic group, at a 5% significance level. The second hypothesis which forced Nanoscypha and Phillipsia species to be monophyletic, thus excluding R. edulis, could be marginally rejected with a variance of +/- 0.41 at a 5% significance level. Hypothesis tree #3, which forced Phillipsia species and R. edulis to be monophyletic, was not rejected by the AU test.
Table 1.
Results of hypothesis testing as determined by Approximately Unbiased (AU, Shimodaira 2002) tests. P-values presented are corrected by Alkaike weights where values greater than 5% (with astericks) indicate tree toologies that are not significantly different from the best tree.

Discussion
In the present study a clade is identified that includes species that have been referred to as the genera Phillipsia, Nanoscypha and Rickiella (Fig. 1). This group, which has reasonable support (Fig. 1), is characterized by moderate to large ascomata, and generally ellipsoid to alantoid spores which are often asymmetrical or flattened on one side. Ascospores are smooth or marked with longitudinal ribs. Within this clade, relationships among the taxa could not be resolved indicating that more data is needed to clarify how the Rickiella, Phillipsia and Nanoscypha lineages are related.AU tests support the lack of monophyly of the genus Phillipsia (Table 1). Clearly species of Phillipsia need additional taxonomic attention. The ability to reject tree #2, which excludes Rickiella from a monophyletic clade with Phillipsia and Nanoscypha species, but not reject tree #3, which considers Rickiella and Phillipsia monophyletic and excludes Nanoscypha, suggests that Rickiella, but not Nanoscypha, may be embedded within a group of Phillipsia species (Table 1). It is interesting to note that Rickiella and Nanoscypha each had unique regions in ITS1 which could not be aligned with each other or with any of the unique regions in this ITS1 hypervariable region described for Phillipsia species by Hansen et al. (1999).
On molecular grounds, as far as studied, there is no justification for accepting or rejecting the genera Rickiella and Nanoscypha but other characters must be considered in determining generic boundaries. There is considerable morphological variation within the clade. Pigments vary from yellow, pink, scarlet, purple to dark greenish. The construction and cell arrangement of the excipular tissues also prove to be diverse. Phillipsia species and R. edulis have an outer excipulum composed of a generally thin prosenchymatous layer as seen in median section. Nanoscypha species have an outer layer that is composed of angular to globose cells. Rickiella edulis is the most distinct morphologically in its unique regularly lacunose excipular tissue (Fig. 2). This feature is unknown in other members of the clade. Given these morphological differences we recognize Rickiella as a genus distinct from Phillipsia. Anamorphic states might prove helpful. Some Phillipsia species, Nanoscypha tetraspora and several other taxa in the Sarcoscyphaceae produce anamorphic states that are placed in the form genus Molliardomyces (Paden 1984, Pfister 1973) but we do not have information on an anamorphic state in Rickiella.

Fig. 2. Macroscopic characters of fresh specimens of Rickiella edulis a) lateral view, b) detail of the lacunose excipular tissue, c) upper view and d) habit of the species as could be founded in the field. Photo credits G. Robledo.
Species of Phillipsia are diverse; a constellation of species or species complexes center on P. domingensis. These fungi produce large ascomata, up to 10 cm diam, have thick flesh, and have spores with longitudinal ridges. ITS sequence data (Hansen et al. 1999) showed little variation within the P. domingensis group and all isolates within this group shared an identical 32 bp in the ITS1 hypervariable region (Hansen et al. 1999). Two taxa, P. carnicolor and P. crispata, differ from the P. domingensis group in their smaller ascomatal size and spore ornamentation. P. crispata and P. carnicolor each have a unique DNA sequence in the ITS1 hypervariable region (Hansen et al 1999). These species produce apothecia up to 2.5 cm diam and have ascospores that are smooth or have very fine longitudinal striate. Phillipsia olivacea may be morphologically distinct as well. In this species ascomata are large (up to about 3.5 cm diam) and have dark green hymenial pigments and smooth or indistinctly striate ascospores that often are nearly alantoid. Furthermore, P. olivacea has a 38 bp sequence in the ITS1 hypervariable region that can be aligned with P. carnicolor but is distinct from the unique DNA sequences (found at this ITS1 position) in the other species in this clade. Both Rifai (1968) and Moravec (1997) noted some of these morphological distinctions within the genus Phillipsia and suggested that subgeneric or generic recognition of them might be appropriate. This view is supported given the DNA sequence differences between species in the ITS1 hypervariable region.
Given the lack of support with the markers we have thus far sampled we are hesitant to break apart Phillipsia but at the same time we are confident in the morphological characters that we have outlined here to support the recognition of both Nanoscypha and Rickiella. Since its introduction Nanoscypha has been accepted by all workers who point to the morphologically distinct excipular construction in this species as a critical character.
In this study the monophyly of the Sarcoscyphaceae is confirmed and Wynnea, which has been placed both in this family and in the Sarcosomataceae, is shown to fall within this well circumscribed family. Our molecular data show three groups (Fig. 1). Two of these groups correspond to the tribes Sarcoscypheae and Boedinopezizeae, as discussed and proposed originally by Korf (1970). Wynnea falls outside both of these tribes and deserves its own tribe. We here provide a diagnosis for a new tribe in the family Sarcoscyphaceae to accommodate Wynnea alone.
Wynneae Pfister, tribe nov.
MycoBank no. 564069
Ascis ut in familia. Ascomis proceris, spathulatis, quarum aliquae observor glomeratae dense in stipite communi, generaliter surgente a sclerotio sepulto. Ascosporis cum cristis longitudinalibus notoriis CB-.
Type genus: Wynnea Berk. & M. A. Curtis, J. Linn. Soc. Bot. 9: 424. 1867.
For literature on this genus see Pfister (1979) and Zhuang (2004).
Rickiella ecology and distribution
Collections of Rickiella edulis are known only from three areas in South America. The lack of collections is striking given the particular morphology of the ascomata. Other than the collection from northwestern Argentina, the species is known from a few examples collected between 1880 and 1922 in Guarapí, Paraguay, Paraguarí Department, the type locality of Peziza edulis Speg., and Rio Grande do Sul state in southeastern Brazil, the type locality of R. transiens (Pfister 1987). Traditional biogeographical summaries place these three areas in three different phytogeographic provinces/regions (Fig.3) within the Neotropical Region (Cabrera and Willink 1973, Morrone 2001). The locality in Paraguay corresponds to Gran Chaco; localities in Rio Grande do Sul correspond to Atlantic Rain Forest/Paranaense Forest; and the northeastern Argentina site corresponds to Yungas Mountain Rain Forests. These three phytogeographic regions show different floristic elements of different origins and, even though the three areas are more or less at similar latitudes, there are different climatic characteristics and altitudes. For example, the area in Argentina where R. edulis was collected has snow at least once a year. In recent biogeographic contributions a new phytogeographic region, the Dry Seasonal Neotropical Forests (DSNF), has been proposed based on distributions of particular tree species and tree species assemblages (Prado 2000, Pennington et al. 2000). Rickiella edulis collections come from the DSNF (Fig.3). In this scenario, it could be supposed that either R. edulis is potentially a widely spread species that is extremely rare or that it has been overlooked, perhaps because these areas have not been sufficiently sampled. We know nothing about its requirements for ascomata production; ascomata may occur only rarely, during a brief season or may be particularly ephemeral. In northwestern Argentina, the species was found growing on dead fallen logs, but the type specimen was said to occur"sur la terre." No particular reference to ecology was reported for collections from southern east Brazil. Collections from Argentina seem to occur on soil (Fig. 2d) but they grew on fallen logs, some of which were partly buried. It is probably the case that the type collection was growing in this way and was misinterpreted as growing on soil. Being a wood-inhabiting species, like nearly all member of the Sarcoscyphaceae, it is plausible that R. edulis follows the distribution of some woody plant of the DSNF. At this juncture such a suggestion is speculative given how little information is at hand. Fungal distribution patterns have received little attention and such distinctive fungi as R. edulis might be helpful in taking an integrated approach to plant and fungal distributions.
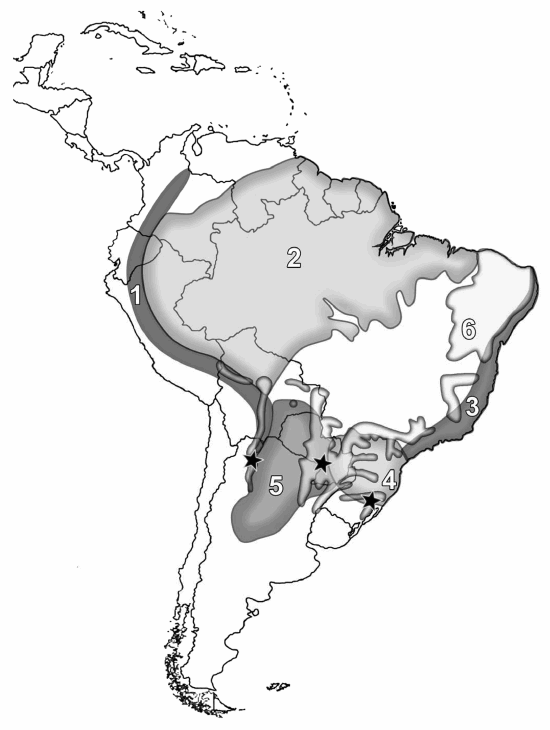
Fig. 3. Known distribution of Rickiella edulis (![]() black stars). Phytogeographic regions in South America are numbered as follows: 1) Yungas Mountain rain forests, 2) Amazonian forest, 3) Atlantic rain forest, 4) Paranaense forests, 5) Gran Chaco and 6) Dry seasonal neotropical forests.
black stars). Phytogeographic regions in South America are numbered as follows: 1) Yungas Mountain rain forests, 2) Amazonian forest, 3) Atlantic rain forest, 4) Paranaense forests, 5) Gran Chaco and 6) Dry seasonal neotropical forests.
Acknowledgements
The authors wish to thank Brad Ruhfel for help with the Approximately Unbiased tree topology testing. We also appreciate the help of Young-Joon Choi on some aspects of the phylogenetic analysis, and Hanno Schaefer and Michaela Schmull for their comments on the manuscript. Karen Hansen provided useful comments on the manuscript, and also gave permission to use previously extracted DNAs. G. Robledo received research support from the National Research Council of Argentina through PIP 6195 to M. Rajchenberg. He kindly acknowledges Idea Wild for their support with technical equipment and is grateful to G. Bertone and A. Bringas (CPA CONICET-UNC) for their technical support. Lic. F. Gelonch (Col. Nac. Monserrat-UNC) is kindly acknowledged for his improvements to the Latin diagnosis. This is publication N° 186 of the PRHIDEB- PROPLAME-CONICET (in respect to A. I. Romero).
References
1. Cabrera A. L. & A. Willink. 1973. Biogeografía de América Latina. Monografías de la O.E.A. Serie Biología. Nro 13. Washington, D.C. [ Links ]
2. Gardes M. & T.D. Bruns. 1993. ITS primers with enhanced specificity for basidiomycets- application to the identification of mycorrhizae and rusts. Mol. Ecol. 2: 113-118. [ Links ]
3. Hansen K., D. H. Pfister & D. S. Hibbett. 1999. Phylogentic relationships among species of Phillipsia inferred from molecular and morphological data. Mycologia 91: 299-314. [ Links ]
4. Hansen K, K. F. LoBuglio & D. H. Pfister. 2005. Evolutionary relationships of the cup-fungus genus Peziza and Pezizaceae inferred from multiple nuclear genes: RPB2, ß-tubulin, and LSU rDNA. Mol. Phylogenet. Evol. 36:1-23. [ Links ]
5. Hansen, K. & D. H. Pfister. 2006. Systematics of the Pezizomycetes-the operculate discomycetes. Mycologia 98: 1029-1040. [ Links ]
6. Harrington, F. A., D. H. Pfister, D. Potter & M.J. Donoghue. 1999. Phylogenetic studies within the Pezizales. I. 18S rDNA sequence data and classification. Mycologia 91: 41-50. [ Links ]
7. Korf, R.P. 1970. Nomenclatural notes. VII. Family and tribe names in the Sarcoscyphineae (Discomycetes) and a new taxonomic disposition of the genera. Taxon 19: 782-786. [ Links ]
8. Korf, R.P. 1983. Cyttaria (Cyttariales): Coevolution with Nothofagus and evolutionary relationship to the Boedijnopezizeae (Pezizales, Sarcoscyphaceae). Aust. J. Bot. Supppl. 10:77-87. [ Links ]
9. Landvik, S., N. F. J. Shailer & O. E. Eriksson. 1996. SSU rDNA sequence support for a close relationship between the Elaphomycetales and the Eurotiales and Onygenales. Mycoscience 37: 237-241. [ Links ]
10. LoBuglio, K. F., J.I. Pitt, & J.W. Taylor. 1993. Phylogenetic analysis of two ribosomal DNA regions indicates multiple independent losses of a sexual Talaromyces state among asexual Penicillium species in subgenus Biverticillium. Mycologia 85 (4): 592-604. [ Links ]
11. LoBuglio, K. F. & D. H. Pfister. 2010. Placement of Medeolariafarlowii in the Leotiomycetes, comments on sampling within the class. Mycol. Progress 9: 361-368. [ Links ]
12. Maddison, D. R. & W. P. Maddison. 2002. MacClade 4.05. Analysis of phylogeny and character evolution. Version 4.0. Sinauer Associates, Sunderland, MA. [ Links ]
13. Mathews, S., M. D. Clements & M. A. Beilstein. 2010. A duplicate gene rooting of seed plants and the phylogenetic position of flowering plants. Phil. Trans. R. Soc. B. 365: 383-395. [ Links ]
14. Miller, M. A., M. T. Holder, R. Vos, P. E. Midford, T. Liebowitz, L. Chan, P. Hoover, T. Warnow. The CIPRES Portals. CIPRES. URL: http://www.phylo.org/sub_sections/portal. 2009-08-04. (Archived by WebCite(r) at http://www.webcitation.org/5imQlJeQa). [ Links ]
15. Moravec, J. 1997. Discomycetes of Madagascar-I. Phillipsiaranomafanensis sp. nov.andascospore sculpture of Cookeina colensoi proved by SEM (Discomycetes, Pezizales, Sarcoscyphaceae). Czech. Mycol. 50: 21-33. [ Links ]
16. Morrone, J. J. 2001.Homology, biogeography and areas of endemism. Diversity and Distributions 7: 297-300. [ Links ]
17. Paden, J. W. 1984. A new genus of Hyphomycetes with teleomorphs in the Sarcoscyphaceae (Pezizales, Sarcoscyphineae). Can. J. Bot. 62: 211-218. [ Links ]
18. Pennington, R. T., D. F. Prado & C. A. Pendry. 2000. Neotrpical seasonally dry forests and quaternary vegetation changes. J. Biogeogr. 27: 261-273. [ Links ]
19. Perry, B. A., Hansen, K. & D. H. Pfister. 2007. A phylogenetic overview of the family Pyronemataceae (Ascomycota, Pezizales). Mycol. Research 111: 549- 571. [ Links ]
20. Pfister, D. H. 1973.Notes on Caribbean discomycetes. III. Ascospore germination and growth in culture of Nanoscypha tetraspora (Pezizales, Sarcoscyphineae). Mycologia 65: 952-956. [ Links ]
21. Pfister D. H. 1979. A monograph of the genus Wynnea (Pezizales, Sarcoscyphaceae). Mycologia 71: 144-159. [ Links ]
22. Pfister, D. H. 1987.The placement of Pezizaedulis in Rickiella (Sarcoscyphaceae, Pezizales). Mycotaxon 29: 329-333. [ Links ]
23. Pfister, D. H., C. Slater, K. Hansen. 2008. Chorioactidaceae: a new family in the Pezizales (Ascomycota) with four genera. Mycol. Research 112: 513-527. [ Links ]
24. Prado, D. E. 2000. Seasonally dry forests of tropical South America: From forgotten ecosystems to a new phytogeographic unit. Edinb. J. Bot. 57: 437-461. [ Links ]
25. Rambaut, A. 1996.Se-AL. Sequence alignment editor.Version 1.0 alpha.University of Oxford, UK. http://evolve.zoo.ox.ac.uk/Se-AL/Se-AL.html. [ Links ]
26. Rifai, M. A. 1968.The Australasian Pezizales in the Herbarium of the Royal Botanic Gardens Kew.Verh. Kon. Ned. Akad. Wetensch., Afd. Natuurk.,Tweede Sect. 57: 1-295. [ Links ]
27. Ronquist, F. & J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572-1574. [ Links ]
28. Ruhfel, B., S. Lindsay & C. C. Davis. 2008. Phylogenetic placement of Rheopteris and the polyphyly of Monogramma (Pteridaceaes.l.): Evidence from Rbcl sequence data. Systematic Botany 33: 37-43. [ Links ]
29. Shimodaira, H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51: 492-508. [ Links ]
30. Swofford, D. L. 2002. PAUP*: Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland, Massachusetts: Sinauer Associates. [ Links ]
31. Thiers, B. [continuously updated]. Index Herbariorum: A global directory of public herbaria and associated staff. New York Botanical Garden's Virtual Herbarium.http://sweetgum.nybg.org/ih/ August 09, 2011. [ Links ]
32. White, T., J. T., Bruns, S. Lee & J. W. Taylor.1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. Pp. 315-322 in PCR Protocols: A Guide to Methods and Applications,eds. MA Innis, DH Gelfard, JJ Sninsky and TJ White, eds. San Diego: Academic Press. [ Links ]
33. Zhuang, W.-Y. 2004. Additional notes on Wynnea (Ascomycetes, Pezizales) from Asia. Nova Hedwigia 79: 519-526. [ Links ]
Original recibido el 1 de Septiembre de 2011;
primera decisión: 1 de Noviembre de 2011;
aceptado el 11 de Noviembre de 2011.
Editor responsable: Carlos Urcelay.














