Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO  uBio
uBio
Compartilhar
BAG. Journal of basic and applied genetics
versão On-line ISSN 1852-6233
BAG, J. basic appl. genet. v.20 n.2 Ciudad Autónoma de Buenos Aires jul./dez. 2009
Is there genetic variation in seedless Argentinean grapefruit? Implications for crop production and conservation
Chacoff, N P1,2*, C P Souto3, M.A. Aizen3 & A Premoli3
1 Laboratorio de Investigaciones Ecológicas de las Yungas, Universidad Nacional de Tucumán. CC 34. Yerba Buena, (4107) Tucumán, Argentina.
2 Actual address: Instituto Argentino de Investigaciones de las Zonas Aridas. CONICET, CC 507, (5500) Mendoza, Argentina. Tel: +54-261-524-4107. Fax: +54-261-524-4001.
3 Laboratorio Ecotono, INIBIOMA-Universidad Nacional del Comahue- CONICET, Quintral 1250, Bariloche (8400) Río Negro, Argentina.
* Corresponding author: Natacha P. Chacoff. E-mail: nchacoff@mendoza-conicet.gov.ar
Título Abreviado: Genetic variation in Argentinean grapefruit
Abstract
Increasing crop genetic diversity has the potential to enhance pollination services, by contributing to the long-term stability of agroecosystems. Different species of the genus Citrus, including grapefruit, are cultivated worldwide. In Argentina, 30% of the total national yield is concentrated in the northwestern part of the country where new cultivars have arisen. We analyzed genetic variation and diversity within and among four plantations of Citrus paradisi for 11 isozyme loci. Although representing single cultivars clonally propagated, within plantations genetic variation was high. Observed heterozygosity ranged from 33 to 36.5%. The number of alleles per locus ranged from 2.4 to 2.6, and polymorphism from 72.7 to 81.8% and for the species in the area reaches 100%. Most of the variation was found within (90%) rather than between plantations. Also, divergence among plantations was only 12%. Despite self-compatibility, fruit production in these plantations benefits from cross-pollination. Therefore, our results highlight the importance of preserving high levels of genetic variation to increase sustainability and productivity of agricultural systems.
Palabras clave: Citrus; Isozymes; Genetic variation; Upper Bermejo River Basin
Resumen
El aumento de la diversidad genética en cultivos posee el potencial de incrementar los servicios de la polinización, contribuyendo, a largo plazo con la estabilidad de los agroecosistemas. Diferentes especies del género Citrus, incluyendo el pomelo, son cultivadas a nivel mundial. En Argentina, el 30% del total de la producción nacional se concentra en la región noroeste del país donde han surgido nuevos cultivares. Analizamos los niveles de variación y diversidad genética dentro y entre cuatro plantaciones de Citrus paradisi para 11 loci isoezimáticos. La variación genética observada dentro de las plantaciones fue alta, a pesar de representar cultivares individuales propagados clonalmente. La heterocigosis observada estuvo entre el 33 y el 36.5%. El número de alelos por locus varió entre 2.4 y 2.6, y el polimorfismo entre 72.7 y 81.8%, mientras que para la especie en el área alcanza el 100%. La mayor parte de la variación observada se encuentra dentro (90%) más que entre las plantaciones. Además, la divergencia entre las plantaciones fue sólo del 12%. Aunque ésta es una especie auto-compatible, la producción de frutos se beneficia con la polinización cruzada. Así, nuestros resultados resaltan la importancia de preservar altos niveles de variación genética para incrementar la sustentabilidad y la productividad de los sistemas agrícolas.
Introduction
Increasing crop genetic diversity has the potential to enhance pollination services, by contributing to the long-term stability of agroecosystems (Hajjar et al., 2008). Currently, there is a global attention on crop yields and productivity in relation to pollinators and the service they provide (Klein et al., 2007, Aizen et al., 2009, Gallaia et al., 2009). These studies highlighted the importance of a diverse and abundant assemblage of pollinators in order to ensure crop production of a wide variety of crops. Many plant species has pollen limitation, which is the deficit of either pollen quantity and/or quality (Ashman et al., 2004, Knight et al., 2005, Aizen and Harder, 2007), and has been recently reported for crops (Chacoff et al., 2008, Vaissiere et al., 2009). This limitation of pollen, has been shown by an increase in fruit production that resulted from cross pollinations compared to self pollination, and has been reported for coffee (Klein et al., 2003), and grapefruit (Chacoff et al., 2008), this result could be due to the presence of within plantations genetic variation. Nevertheless, is striking the lack of surveys analyzing within crop genetic diversity.
In natural populations, the amount of extant genetic variation may influence their short-term viability and their long-term potential for evolutionary change (Masatoshi, 1987, Hamrick and Godt, 1996) and in plantations their yield and economic profitability (Torres et al., 1978, Escalante et al., 1994, Roubik, 2002). Although much information exists on the amounts and partitioning of genetic variation in naturally growing (Hamrick and Godt, 1989) and cultivated plant species (Hamrick and Godt, 1997), less is known on how this variation is partitioned within and among cultivars. Citrus is one of the most economically important genus among cultivated fruit trees, growing in tropical and subtropical regions and cultivated over a wide range of latitude. Citrus is almost universally propagated by budding scion cultivars onto apomictic seedling rootstocks. This leads to the clear expectation that, within a plantation, there will be a high level of genetic uniformity. This has been observed in many studies using isozymes and DNA markers (Fang and Roose, 1997, Elisiario et al., 1999). Furthermore, most "grapefruit" cultivars have been developed by selection of either apomictic seedlings or, more commonly, bud sport mutations (Frost and Soost, 1968). Cultivars that originate in this way are expected to be nearly identical for genetic markers. Many studies have also reached this conclusion (e.g. Torres et al., 1978).
The hybrid origin of grapefruit is well documented. Species that arose by hybridization between other taxa may have elevated heterozygosity, especially if these species are highly apomictic (Federici et al., 1998). This was the case of C. paradisi where RFLP and RAPD analyses revealed one of the highest heterozygosity indices among 88 accessions representing 45 Citrus species (Federici et al., 1998). Also large amounts of intraspecific variation exist in leaf, plant, and fruit characters of this species. Nevertheless, molecular analyses provide complementary data to morphological information for plant germoplasm classification (Tinker et al., 1994, Morgante et al., 1996), and different molecular markers have been used to distinguish genetic diversity in grapefruit (Torres et al., 1978, Kijas et al., 1995, Fang and Roose, 1997). In particular, Torres et al. (1978) proposed that isozymes provide excellent single-gene markers because they are colinear, codominant, little affected by the environment, and are identical in leaves of young and mature plants of the same cultivated variety. Although, more recently, studies on allelic expression variation attributed to differences in noncoding DNA sequences, play with that variation in different environments along plant maturation to establish their role in determining phenotypic diversity (Guo et al. 2004).
In NW Argentina, grapefruit industry is economically important, and new cultivars have been developed in situ. Pollination biology of this cultivars revealed that an insect vector is needed to set fruits and that, although this cultivars are self compatible, exists a relative advantage in pollen germination from cross relative to self pollen (Chacoff and Aizen, 2007). Moreover, hand pollinated flowers with pollen from different plants within the same cultivar showed a relative and consistent increment in fruit set than hand pollinated flowers within flowers of the same tree (Chacoff et al., 2008). These results suggest that these grapefruit plantations are not genetically uniform. We used isozyme electrophoresis to assess genetic diversity within and among four grapefruit plantations, from three different cultivars, two of them developed within the region.
Materials and Methods
This study was conducted in grapefruit plantations near Orán city in Salta province, Argentina. These plantations are located in the Upper Bermejo River Basin within the premontane lowlands forest known as Yungas (23°28' S; 64°24'W). In this area, Citrus plantations occupy 13.500 ha, most of them are sweet orange (32%) and grapefruit (57%). Grapefruit plantations of this area represent more than 30% of the national production; that is mostly (>80%) exported (Dansa, 2001). Grapefruit cultivars in this area are mainly from red and very red seedless cultivars (Henninger's ruby, Río Red, Rouge La Toma, Flame, Foster seedless and Star Ruby) (Peralta, 1999).
Grapefruit cultivars and plantations:
We studied three seedless pigmented cultivars in four plantations. Trees in the studied plantations are clonally propagated on rootstocks as nearly all commercial citrus trees are in most parts of the world (Gmitter, 1995). The cultivars were:
1. 'Rouge La Toma' is a very red seedless cultivar that was studied at La Toma SA and Produnor SA. This is a local variety originated in La Toma SA plantation from a natural mutation that occurred in a Henninger's ruby tree that was selected and propagated asexually (Ing. R. Burgos, personal communication). This cultivar is widespread in the area (Peralta, 1999, Dansa, 2001, Federcitrus, 2003). Trees in this plantation had approximately 15 years.
2. 'Rio Red' is a very red seedless cultivar that was studied in the plantation of Citrusalta SA. This variety originated in 1963 in Texas (USA). Buds of Ruby red seedlings trees were irradiated with thermal neutrons; the new clone was selected after the appearance of a sport mutation (Gmitter, 1995). Plantation had 9 years approximately.
3. 'Foster seedless' is a red seedless grapefruit that was studied in Pena Colorada. This cultivar arose from a natural mutation in a Foster tree in the Institute of Agronomic Technology (INTA) at Yuto, Jujuy province in Argentina (Ing. J. Palacios, personal communication). This mutation was selected and propagated as Foster seedless and is intensively cultivated in the area (Peralta, 1999). Plants within this plantation had 5 years mixed with some of 40 years that were replaced by younger ones.
The origin of all of these plantations is by vegetative shoots, which comes from a set of selected trees (Personal communication with the owners of the plantations).
Field sampling and laboratory work:
Fresh leaf material was collected from a total of 260 plants that were randomly chosen from each plantation (68 from Citrusalta SA, 67 from Produnor SA, 59 from La Toma SA, and 66 from Pena Colorada). Samples were kept fresh and transported in a portable cooler to the laboratory where enzymes were extracted using the buffer of Mitton et al. (1979). Homogenates were stored at -80oC until horizontal electrophoresis on 12% w/v starch gels was conducted. We determined the genotype of all collected individuals at different isozyme loci. The loci were considered putative, as we did not carry out formal genetic analysis in C. paradisi. However, the observed banding patterns were typical of the same enzymatic systems reported in species for which formal analysis has been conducted (e.g. Soltis et al., 1983).
We resolved nine enzyme systems that coded for eleven loci using the Tris-citrate electrobuffer and the Histidine - EDTA gel buffer, pH 7 (King and Dancik, 1983). Isozyme loci were isocitrate dehydrogenase (IDH), malate dehydrogenase (MDH), malic enzyme (ME), menadione reductase (MNR-1, MNR-2), 6-phosphogluconate dehydrogenase (6PGD), peroxidase (PRX cathodal and anodal), phosphogluco isomerase (PGI), phosphogluco mutase (PGM), and shikimate dehydrogenase (SKDH). Electrophoresis was carried out at 4°C for 6 hours at 30mA until the bromophenol blue marker had moved 10 cm from the origin towards the anode. The anodic and cathodic portions of the gels were sliced horizontally in four sections, each 1 mm thick. Stain procedures for the studied enzymes followed commonly used protocols reported elsewhere (Conkle et al., 1982, Soltis et al., 1983). The alleles were numbered sequentially, with the lowest number indicating the fastest moving towards the anode.
Data Analysis:
Levels of isozyme variation at plantation level were described by standard gene-diversity measures using POPGENE v.1.31 (Yeh et al., 1997). These were the mean number of alleles per locus (A), the observed heterozygosity (HO), and the percentage of polymorphic loci (P) using the 95% criterion. Genetic diversity was analyzed following Nei (1973) by the total (HT) and within-population (HS) diversity. Differentiation among plantations was measured by F statistics. We estimated within-plantation fixation with Fis and among-plantation divergence by Fst (Wright, 1965) using FSTAT v. 2.9.1 (Goudet, 2000). Mean and 95% confidence intervals (CI) were calculated by jackknifing and bootstrapping over polymorphic loci, respectively. These parameters were estimated for all loci since all were polymorphic. Deviations of Fis from zero were assessed using chi-square tests. We used analysis of molecular variance (AMOVA) to estimate variance components for isozyme phenotypes, partitioning the variation among individuals/within plantations and among plantations (Excoffier L, 1992). This analysis was undertaken with GenAlEx (Peakall and Smouse, 2006).
Results
All putative isozyme loci scored in Citrus paradisi (11/11) were polymorphic in at least one plantation (Table I). We detected a total of 34 different alleles, 22 were present in all plantations. Four alleles were exclusive of Produnor (6Pgd-1, Idh-3, Mnr1-4, Prx-an-3); and one of Pena Colorada (Prx-an-1) (Table I). The observed amount of genetic variation is high although, differences among plantations were moderate (Table II). On average all plantations have 2.6 alleles per locus and, observed heterozygosity was 0.34. Two plantations have 72.7% and the other two 81.8% of polymorphism loci (Table II). In addition, we found substantial amounts of total and within-plantation genetic diversity (HT= 0.375 and HS = 0.341, respectively).
Table I: Allelic frequencies for each of the 4 plantations of Citrus paradisi of NW Argentina.
| Plantation | |||||
| Locus | Allele # | Produnor | Pena Colorada | Citrusalta | La Toma |
| 6Pgd | 1 | 0.02 | 0 | 0 | 0 |
| 2 | 0.55 | 0.26 | 0.18 | 0.08 | |
| 3 | 0.43 | 0.74 | 0.81 | 0.88 | |
| 4 | 0 | 0 | 0.01 | 0.04 | |
| Idh | 1 | 0.02 | 0.08 | 0 | 0.06 |
| 2 | 0.85 | 0.92 | 1 | 0.94 | |
| 3 | 0.13 | 0 | 0 | 0 | |
| Mdh | 1 | 0.01 | 0.02 | 0 | 0.04 |
|
| 2 | 0.06 | 0.05 | 0.09 | 0.28 |
|
| 3 | 0.24 | 0.15 | 0.3 | 0.30 |
|
| 4 | 0.69 | 0.78 | 0.61 | 0.38 |
| Me | 1 | 0.79 | 0.67 | 0.81 | 0.97 |
| 2 | 0.21 | 0.33 | 0.19 | 0.03 | |
| Mnr-1 | 1 | 0 | 0 | 0.04 | 0.06 |
| 2 | 0.41 | 0.52 | 0.44 | 0.43 | |
| 3 | 0.58 | 0.48 | 0.52 | 0.51 | |
| 4 | 0.01 | 0 | 0 | 0 | |
| Mnr-2 | 1 | 0.98 | 1 | 0.98 | 1 |
| 2 | 0.02 | 0 | 0.02 | 0 | |
| Pgi | 1 | 0 | 0.04 | 0 | 0.01 |
| 2 | 0.78 | 0.73 | 0.81 | 0.45 | |
| 3 | 0.22 | 0.23 | 0.19 | 0.54 | |
| Pgm | 1 | 0 | 0.04 | 0.07 | 0.04 |
| 2 | 0.05 | 0.3 | 0.56 | 0.56 | |
| 3 | 0.94 | 0.4 | 0.29 | 0.34 | |
| 4 | 0.01 | 0.26 | 0.08 | 0.06 | |
| Prx-an | 1 | 0 | 0.02 | 0 | 0 |
| 2 | 0.98 | 0.98 | 1 | 1 | |
| 3 | 0.02 | 0 | 0 | 0 | |
| Prx-cat | 1 | 0.65 | 0.6 | 0.61 | 0.40 |
| 2 | 0.35 | 0.4 | 0.39 | 0.60 | |
| Skdh | 1 | 0.25 | 0.07 | 0.15 | 0.04 |
| 2 | 0.44 | 0.5 | 0.58 | 0.73 | |
| 3 | 0.31 | 0.43 | 0.27 | 0.23 | |
Table II: Indexes of genetic variability in 11 loci of Citrus paradisi from four plantations from the NW of Argentina. Mean number of alleles per locus (A) and standard error (SE); Mean observed heterozygosity (Ho) standard error (SE); and P95%.
| Plantation | Cultivar (variety) | A (S.E) | Ho (S.E) | P95% |
| Pena Colorada | Foster Seedless (Red) | 2.45 (0.28) | 0.37 (0.07) | 81.82 |
| CitruSalta | Rio Red (Very red) | 2.36 (0.28) | 0.33 (0.07) | 72.73 |
| Produnor | Rouge La Toma (Very red) | 2.64 (0.20) | 0.34 (0.06) | 81.82 |
| La Toma | Rouge La Toma (Very red) | 2.55 (0.31) | 0.33 (0.08) | 72.73 |
| Average | 2.5 (0.27) | 0.34 (0.07) | 77.27 |
Estimates of Fis (means ± SE) by jackknifing over loci were 0.022 ± 0.176. The 95% confidence intervals for Fis were [-0.267, 0.334], indicating that, on average, the observed number of heterozygotes did not differ significantly from expectation. However, fixation indices were significantly different from zero in 27 of 39 (69%) possible estimates, 59% of which (16 of 27) yielded a positive deviation from zero, indicating heterozygote deficiencies at many loci (Table III). In addition, we found significant genetic differences among plantations (Fst =12%; CI95% = [0.052, 0.198]).
Table III: Fixation indexes (Fis) at 11 isozyme loci in 4 plantations of Citrus paradisi of NW Argentina.
| Loci | Produnor | Pena Colorada | Citrusalta | La Toma |
| 6Pgd | -0.593* | 0.376* | -0.092 | -0.105 |
| Idh | -0.157 | -0.087 | NA | 1.000* |
| Mdh-1 | 0.216* | 0.399* | 0.547* | NA |
| Me | -0.273 | -0.493* | -0.235 | -0.032 |
| Mnr-1 | -0.690* | -0.843* | -0.796* | -0.700* |
| Mnr-2 | 1.000* | NA | -0.020 | 0.672* |
| Pgi-1 | 0.531* | 0.564* | 0.285* | 0.395* |
| Pgm-1 | 0.648* | 0.412* | 0.628* | 0.567* |
| Prx-an | 1.000* | -0.020 | NA | NA |
| Prx-ca | -0.081 | 0.083 | -0.387* | 0.106 |
| Skdh | -0.510* | -0.678* | -0.373* | -0.150* |
| Average Fis | 0.052 | 0.125 | 0.028 | 0.181 |
Note: NA is not aplicable. *P<0.05 Statistically different from 0 in Chi-square tests.
The AMOVA, showed that only 10% of the observed differences could be attributable to among plantation genetic variation and that most of the observed variation is among individuals/within plantations (Fig. 1 and Table IV).
Figure 1: Percentage of genetic variation found among and within four grapefruit plantations from NW of Argentina
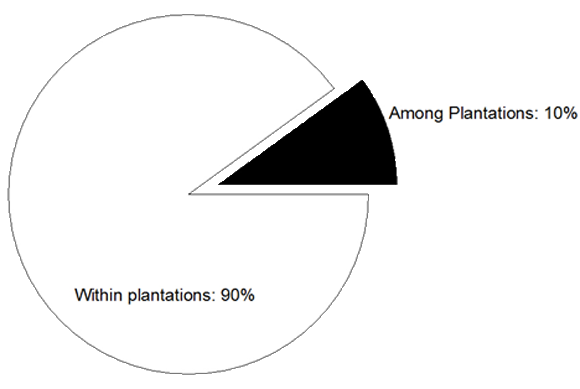

Table IV: Results of the analysis of molecular variance (AMOVA) of genetic variation for Citrus paradisi
| Source of Variation | Df | SS | MS | P |
| Among Plantations | 3 | 66.|43 | 22.048 | |
| Within Plantations | 392 | 742.349 | 11.894 | 0.010 |
Df: degree of freedom. SS: sum of squares. MS: mean Squire. P: statistical probability.
Discussion
We achieved good activity and separation in Citrus paradisi for all studied enzymatic systems including ones previously reported as poorly resolved (Torres et al., 1978, Novelli et al., 2000, Cabrita et al., 2001). The observed levels of isozyme variation were comparable with the ones reported for Citrus spp. in general (A = 2.5, HO = 0.34, P = 85%) (A = 2.5, HO = 0.34, P = 85%; Novelli et al., 2000). For grapefruit, in particular, Torres et al. (1982) found that 2 of 10 screened genes were polymorphic (Got 1 and Lap). In two C. paradisi hybrids, Novelli et al. (2000) reported that 5 and 8 of 19 loci were polymorphic (PGI2, PGM1, Lap1, Lap2, Aat1, 6PGD2, SKDH1, Acp2), respectively. Our study revealed higher levels of polymorphism as 11 of 11 isozyme loci resulted to be polymorphic in at least one plantation.
Our results showed that Argentine grapefruit plantations are unexpectedly genetically diverse (HT = 0.37) compared with those of other cultivated species, where HT's ranges between 0.14 to 0.23 (Hamrick and Godt, 1989).
The hybrid origin of grapefruit and the occurrence of somatic mutations, including chimeras (Frost and Soost, 1968), could explain the high levels of genetic variation found in NW Argentine grapefruit plantations. Although some authors suggested that isozymes may not be selectively neutral (Brown 1971; Mueller et al. 1985), statistical analysis assumptions considered them as neutral genetic markers. Nevertheless, the observed genetic variation does not necessarily express itself in fruit characteristics, but this variable gene pool represents a reservoir for improvement of grapefruit cultivars. Despite the high levels of polymorphism found in grapefruit plantations from NW Argentina, fixation indices indicate a deficiency of heterozygotes at many loci. This excess of homozygotes may be explained by the clonal propagation of genetically identical individuals within each plantation.
Oddly enough, the high intraplantation or intracultivar genetic variation found in red grapefruit from NW Argentina was much larger than expected for a clonally-propagated plantation, resembling levels of genetic diversity found in natural-outcrossing populations (Hamrick and Godt, 1996). In addition to the occurrence of high rates of somatic mutations, the high levels of genetic variation could be attributed to a number of causes including propagation via monoembryonic seeds, multiple origin of the cultivar material, and even mistaken labeling of seedlings. It is relatively common that different genotypes may be marketed under the same name in the same or different areas.
Isozyme analysis proves useful to determine the origin and clonal nature of a plantation. Ashari et al. (1989) had shown that isozyme techniques can be used successfully not only to discriminate between mandarin types, but also to investigate the parentage and relatedness of the cultivars. It is clear that the success of isozyme analysis increases with the number of enzyme systems employed. The biochemical technology for detecting enzymes in gels is now at a stage where a wide range of isozyme loci can be tested relatively easily. These methods are now readily available to both horticulturalists and plant breeders for routine confirmation of the genotype of their material (Murphy et al., 1996). Also, Elisiario et al. (1999) found that age or environment exerts low influence on the leaf isozyme systems analysis, confirming their suitability for genetic analysis and to fingerprint genotypes in Citrus.
Thus, even when isozyme analysis in this case may not be the most appropriate method for identification and discrimination among cultivars, it is still a useful tool to detect within plantations genetic variation. The locally generated cultivars (i.e. Rouge La Toma and Foster Seedless) present in three out of the four studied plantations have a short history of cultivation, so variation within them is highly expected mainly considering that they are not stable. These cultivars also harbor a number of exclusive alleles. On the other hand, the Rio Red cultivar, planted at Citrusalta is the only variety studied that was not locally originated. This variety has a long history of cultivation, and is the one that showed the lowest levels of polymorphism (Table II). In addition, we found significant amounts of total and within-plantation genetic diversity (HT= 0.375 and HS = 0.341, respectively).
Implications:
The current study report two surprising results, first there are high levels of genetic variation within grapefruit plantations and that almost 90% of the observed differences could be attributable to among plantation genetic variation, evidencing little difference among plantations.
These results open some questions that were unanswered in this study:
a) Does isozyme heterogeneity have any useful corresponding variation in agronomic traits?
b) Does improved fruitset and quality really result from crossing different genotypes, or just from crossing different plants per se?
c) Are spontaneous mutations that promote genetic diversity driven by the fitness advantage of better fruit production from outcrosses?
d) Are genetic changes over time?
e) Is there genetic variation within a single tree?
Most cultivated species are not intended to be genetically diverse. One assumes that if the isozyme diversity discovered here was also reflected in the tree and fruit characteristics, it would be a highly undesirable situation. Despite the small differences among plantations, these cultivars are clearly distinguished by tree and fruit phenotype. Thus, the observed genetic diversity could act as a reservoir for improvement of grapefruit cultivars.
Acknowledgments
We thank to R. Manero, J. Campos, R. Burgos and D. Lorenzo for allowing us to conduct this study in their plantations. Rebecca Lobo, Carolina Monmany, Gabriela Elias, and Valeria Aschero assisted during fieldwork. Alejandro Brown provided logistic assistance during this work. The Antorchas Foundation (Grant 14022-22), Pro-Yungas Foundation, Sigma Xi and the National Research Council of Argentina (CONICET) founded this research. Comments from an anonymous reviewer were helpful to achive a better comprehension of the manuscript.
References
1. Aizen MA, LA Garibaldi, SA Cunningham & AM Klein (2009) How much does agriculture depend on pollinators? Lessons from long-term trends in crop production. Annals of Botany, 103, 1579-1589. [ Links ]
2. Aizen MA & LD Harder (2007) Expanding the limits of the pollen-limitation concept: Effects of pollen quantity and quality. Ecology, 88, 271-281. [ Links ]
3. Ashari S, D Aspinall & M Sedgley (1989) Identification and investigation of relationships of mandarin types using isozyme analysis. Scientia Horticulturae, 40, 305-315. [ Links ]
4. Ashman T, TM Knight, JA Steets, P Amarasekare, M Burd, DR Campbell, MR Dudash, MO Johnson, SJ Mazer, RJ Mitchell, MT Morgan & WG Wilson (2004) Pollen limitation of plant reproduction: Ecological and evolutionary causes and consequenses. Ecology, 85, 2408-2421. [ Links ]
5. Brown AHD (1971) Isozyme Variation under Selection in Zea mays. Nature 232, 570 - 571. [ Links ]
6. Cabrita L, P Elisiario, J Leitao & A Guerreiro (2001) Assessment of the genetic relationship among Citrus species and varieties by isozyme and RAPD markers. Acta Horticulturae, 546, 177-181. [ Links ]
7. Chacoff NP & MA Aizen (2007) Pollination Requirements of Pigmented Grapefruit (Citrus paradisi Macf.) from Northwestern Argentina. Crop Sci, 47, 1143-1150. [ Links ]
8. Chacoff NP, MA Aizen & V Aschero (2008) Proximity to forest edge does not affect crop production despite pollen limitation. Proceedings of the Royal Society B: Biological Sciences, 275, 907-913. [ Links ]
9. Conkle MT, PD Hodgkiss, LB Nunnally & SC Hunter (1982). Starch gel electrophoresis of conifer seeds: a laboratory manual. General Technical report. Berkeley, California, USA: Pacific Southwest Forest and Range Experimental Station, Forest Service, U.S.D.A. [ Links ]
10. Dansa AM (2001). Perfil de mercado de Cítricos. 1-25. Buenos Aires: Secretaria de Agricultura, Ganadería, Pesca y Alimentos. [ Links ]
11. Elisiario PJ, EM Justo & JM Leitao (1999) Identification of mandarin hybrids by isozyme and RAPD analysis. Scientia Horticulturae, 81, 287-299. [ Links ]
12. Escalante AM, G Coello, LE Eguiarte & D Pinero (1994) Genetic structure and mating systems in wild and cultivated populations of Phaseolus coccineus and P. vulgaris (Fabaceae). American Journal of Botany, 81, 1096-1103. [ Links ]
13. Excoffier L SPaQJ (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. . Genetics. , 131, 479-491. [ Links ]
14. Fang DQ & ML Roose (1997) Identification of closely related citrus cultivars with inter-simple sequence repeat markers. Theorical Applied Genetics, 95, 408-417. [ Links ]
15. Federcitrus (2003). Estadísticas 2003. Ranking de Empresas Exportadoras. www.atcitrus.com [ Links ]
16. Federici CT, DQ Fang, RW Scora & ML Roose (1998) Phylogenetic relationships within the genus Citrus (Rutaceae) and related genera as revealed by RFLP and RAPD analysis. Theorical Applied Genetics, 96, 812-822. [ Links ]
17. Frost HB & RK Soost (1968). Seed reproduction: Development of gametes and embryos. In The Citrus Industry, eds). W Reuther, HJ Webber & LD Batchelor, 290-320. California, USA: University of California Press. [ Links ]
18. Gallaia N, J-M Salles, J Setteled & BE Vaissiere (2009) Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecological Economics, 68, 810-821. [ Links ]
19. Gmitter FGJ (1995) Origin, evolution and breeding of the grapefruit. Plant Breeding Reviews, 13, 345-363. [ Links ]
20. Guo M, MA Rupe, C Zinselmeier, J Habben, BA Bowen & OS Smith (2004) Allelic Variation of Gene Expression in Maize Hybrids The Plant Cell 16, 1707-1716 [ Links ]
21. FSTAT. A program to estimate and test gene diversities and fixation indices. release 2.9.1., Université de Lausanne, Dorigny, Switzerland. [ Links ]
22. Hajjar R, DI Jarvis & B Gemmill-Herren (2008) The utility of crop genetic diversity in maintaining ecosystem services. Agriculture, Ecosystems and Environment, 123, 261-270. [ Links ]
23. Hamrick JL & JW Godt (1989). Allozyme diversity in plant species. In Plant Population Genetics, Breeding, and Genetic Resources, eds). AHD Brown, MT Clegg, AL Kahler & BS Weir, 43-63. Sunderland, Massachusetts, USA: Sinauer Associates. [ Links ]
24. Hamrick JL & JW Godt (1996) Effects of life history traits on genetic diversity in plant species. Phil. Trans. R. Soc. Lond. B . 351, 1291-1298. [ Links ]
25. Hamrick JL & MJW Godt (1997) Allozyme diversity in cultivated crops. Crop Science 37, 26-30. [ Links ]
26. Kijas JMH, JCS Fowler & MR Thomas (1995) An evaluation of sequence tagged microsatellite site markers for genetic analysis within citrus and related species. Genome, 38, 349-355. [ Links ]
27. King JN & BP Dancik (1983) Inheritance and linkage of isozymes in white spruce (Picea glauca). Canadian Journal of Genetics and Cytology, 25, 430-436. [ Links ]
28. Klein AM, I Steffan-Dewenter & T Tscharntke (2003) Fruit set of highland coffee increases with the diversity of pollinating bees. Proceedings of the Royal Society of London. B., 270, 955-961. [ Links ]
29. Klein AM, BE Vaissiere, JH Cane, I Steffan-Dewenter, SA Cunningham, C Kremen & T Tscharntke (2007) Importance of pollinators in changing landscapes for world crops. Proceedings of the Royal Society B: Biological Sciences, 274, 303-313. [ Links ]
30. Knight TM, JA Steets, JC Vamosi, SJ Mazer, M Burd, DR Campbell, MR Dudash, MO Johnston, RJ Mitchell & T-L Ashman (2005) Pollen limitation of plant reproduction: Pattern and process. Annual review of Ecology, Evolution, and Systematics, 36, 467-497. [ Links ]
31. Masatoshi N (1987). Genetic variation within population. In Molecular evolutionary genetics, (ed. M Nei, 176-207. New York: Columbia University Press. [ Links ]
32. Mitton JB, YB Linhart, KB Sturgeon & JL Hamrick (1979) Allozyme polymorphisms detected in mature needle tissue of ponderosa pine. Journal of Heredity, 70, 86-86. [ Links ]
33. Morgante M, A Pfeiffer, A Costacurta & AM Olivieri (1996) Molecular tools for population and ecological genetics in coniferous trees. Phyton, 36, 133-142. [ Links ]
34. Mueller LD, LG Barr & FJ Ayala (1985) Natural selection vs. random drift: evidence from temporal variation in allele frequencies in nature. Genetics 111, 517-554. [ Links ]
35. Murphy RW, JWJ Sites, DG Buth & CH Haufler (1996). Proteins: Isozyme electrophoresis. In Molecular Systematics, eds). DM Hillis, C Moritz & BK Mable, 51-120. Sunderland. MA: Sinauer Associates. [ Links ]
36. Nei M (1973) Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences, 70, 3321-3323. [ Links ]
37. Novelli VM, MA Machado & C Romero Lopes (2000) Isoenzymatic polymorphism in Citrus spp. and Poncirus trifoliata (L.) Raf. (Rutaceae). Genetics and Molecular Biology, 23, 163-168. [ Links ]
38. Peakall R & PE Smouse (2006) Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6, 288-295. [ Links ]
39. Peralta C (1999). Informes Citrícolas de la Provincia de Salta. 28-35. INTA ORAN, SALTA. [ Links ]
40. Roubik DW (2002) The values of bees to the coffee harvest. Nature, 417, 708. [ Links ]
41. Soltis DE, CH Haufler, DC Darrow & GJ Gastony (1983). Starch gel electrophoresis of ferns. A compilation of grinding buffers, gel and electrode buffer, and staining schedules. New York, USA.: W. H. Freeman. [ Links ]
42. Tinker NA, DE Mather & MG Fortin (1994) Pooled DNA for linkage analysis: Practical and statistical consideration. Genome, 37, 999-1004. [ Links ]
43. Torres AM, RK Soost & U Diedenhofen (1978) Leaf isozymes as genetic markers in Citrus. American Journal of Botany, 65, 869-881. [ Links ]
44. Torres AM, RK Soost & T Mau-Lastovicka (1982) Citrus isozymes: Genetics and distinguishing nucellar from zygotic seedlings. Journal of Heredity, 75, 335-339. [ Links ]
45. Vaissiere BE, G Carré, J Vandame & B Gemmill-Herren (2009). Detecting and assessing pollination deficit in crops. ed. F REPORT, 63. Avignon, France: Institut National de la Recherche Agronomique (INRA) and Food and Agriculture Organization of the United Nations (FAO). [ Links ]
46. Wright S (1965) The interpretation of population structure by F-Statistics with special regard to systems of mating Evolution, 19, 395-420. [ Links ]
47. POPGENE, the user-friendly shareware for population genetic analysis. University of Alberta, Canada. [ Links ]














