Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
BAG. Journal of basic and applied genetics
On-line version ISSN 1852-6233
BAG, J. basic appl. genet. vol.21 no.1 Ciudad Autónoma de Buenos Aires Jan./June 2010
ARTÍCULOS ORIGINALES
Somaclonal variation in Cereus peruvianus Mill. (Cactaceae): its potential to generate new varieties and broaden the species's genetic basis
Resende Gazoli Adriana, Claudete Aparecida Mangolin, Machado Maria de Fátima Pires da Silva*
Department of Cell Biology and Genetics. State University of Maringá. 87020-900 Maringá PR Brazil.
*Corresponding author: mfpsmachado@uem.br
ABSTRACT
In current study Random Amplified DNA-Polymorphism markers were used to detect genetic variations in C. peruvianus somaclones and to examine relationships among somaclones and plants grown from seeds in cultivated populations. The possible association of DNA fragments with morphological traits of somaclones was also investigated. Polymorphism in somaclones (62.24%) was higher than that detected in cultivated plants (42.57%). The casual grouping of somaclones presented atypical and typical shoot phenotypes showed that the random amplifi ed DNA-fragment polymorphisms detected in present study were not associated with shoot morphologies. Similarity in somaclones and cultivated plants ranged from 70.2 to 93.7% and 76.7 to 86.8%, respectively. Thus, in vitro tissue culture of C. peruvianus may be recommended to broaden the species's genetic base. The genetic variability found in somaclones is important as a source of new traits that may be necessary as breeding or conservation programs proceed.
Key Words: Cactus; Genetic diversity; Mandacaru; RAPD markers; Somaclonal variation.
RESUMO
No presente estudo o polimorfi smo de seqüencias de DNA amplificadas aleatoriamente foram usadas como marcadores moleculares para detectar variação genética em somaclones de Cereus peruvianus e para examinar a relação entre os somaclones e as plantas que crescem a partir de sementes, em populações cultivadas. A possível associação entre fragmentos de DNA com características morfológicas de somaclones também foi investigado. O polimorfi smo nos somaclones (62,24%) foi maior do que o detectado em plantas cultivadas (42,57%). O agrupamento casual dos somaclones apresentando caules com morfologias atípicas e típicas mostrou que o polimorfismo dos fragmentos de DNA amplifi cados no presente estudo não está associado com morfologias de caules. A similaridade nos somaclones e nas plantas cultivadas variou de 70,2 a 93,7% e de 76,7 a 86,8, respectivamente. Portanto, a cultura de tecido in vitro de C. peruvianus pode ser recomendada para ampliar a base genética desta espécie. A variabilidade genética encontrada nos somaclones é importante com uma fonte de novas características que podem ser necessárias para procedimentos em programas de melhoramento ou de conservação da espécie.
Palavras-chaves: Cactos; Diversidade genética; Mandacaru; Marcadores RAPD; Variação somaclonal.
INTRODUCTION
It is now well documented that tissue cultures may induce heritable genetic changes in plants. Larkin and Scowcroft (1981) adopted the term somaclonal variation to describe the genetic variation occurring in in vitro cultured cells and they suggested the potentiality of tissue culture for the induction of useful and stable variations that could be exploited for crop improvement. A considerable number of useful somaclonal variants have been generate and recorded from different plant species (Jain, 2001; Taji et al., 2002). Somaclones of the Cereus peruvianus cactus species, for instance, exhibit considerable isozyme patterns and morphological variation (Mangolin et al., 1997; Machado et al., 2000).
C. peruvianus, popularly known in Brazil as mandacaru, is a columnar cactus species found in arid and semiarid regions in the North and Northwest Brazil. It is also distributed in fragments of subtropical forests in the South and Southwestern regions of Brazil and is a common ornamental cactus cultivated in gardens. According to Nerd et al. (2002) C. peruvianus is under domestication in Israel and is sold in Europe; their fruits were accepted very well in both local and European markets, due to its beautiful appearance, delicate sour-sweet taste, and unique aroma. Self-incompatibility has been reported for C. peruvianus as well as for other night-flowering columnar cactus; fruit was produced by open-pollination and manual crosspollination, but not by self-pollination (Weiss et al., 1993, 1994; Silva and Sazima, 1995; Casas et al., 1999; Ruvolo-Takasusuki et al., 2006). In Brazil, the plants of C. peruvianus have also been used as source of chemical compounds for the cosmetic, food, and pharmaceutical industry (Alvarez et al., 1992, 1995; Nozaki et al., 1993; Barros and Nozaki, 2002; Oliveira and Machado, 2003).
Since C. peruvianus has industrial importance the callus tissue culture was induced in order to obtain a considerable number of regenerated plants (Oliveira et al., 1995). In vitro callus culture of C. peruvianus has been a faster and efficient method for clonal multiplication of plants (Oliveira et al., 1995). Somaclones, also designed clonal regenerants (R0), were regenerated from callus culture induced using MS medium (Murashige and Skoog, 1962) containing B5 vitamins (Gamborg et al., 1968), 0.8% agar, 3% sucrose, and combinations of the growth regulators 2,4-dichlorophenoxyacetic acid (2,4-D) and N-(2-furanylmethyl)-1H-purine-6 amine (kinetin) according to the method of Oliveira et al. (1995). Callus tissue was formed from hypocotyls stalks of three seedlings of C. peruvianus which were obtained from seeds of one naturally cultivated plant. Callus subcultures were performed at 15-day intervals, and 18-20 weeks after culture initiation (total of 23 somaclonal generations) the friable calli produced the cactus shoots (somaclones). The somaclone population (R0 plants) was planted in 1997 at the Experimental Botanic Garden of the State of University of Maringá (Maringá PR Brazil; altitude 554.9m; 23º 25'S; 51º 25' W) where they are still maintained.
Differential morphological types of shoots were reported in 4-year-old plants of C. peruvianus regenerated from callus tissues (Mangolin et al., 1997; Machado et al., 2000). Moreover, at least three types of shoots may be observed in 10-year-old somaclones: i) typical or regular shoots similar to those found in C. peruvianus plants growing from seeds in cultivated populations (62.5%); ii) atypical shoots in which the areoles are found in broken ribs forming alternate knobby regular or irregular space, sometimes with tortuously formed areoles similar to spiraled ribs (23%); and iii) mixed shoots (14.5%) showing mixed morphological forms (typical and atypical parts).
Since the ratio of shoot-morphological variation is high, it is reasonable to suppose that the variant phenotype is caused by genetic and/or epigenetic changes. Consequently, a higher degree of genetic diversity at DNA level may be detected among somaclones than that among C. peruvianus plants grown from seeds in natural populations. Higher degree of genetic diversity among C. peruvianus plants is important because Gutman et al. (2001) have shown that this specie has only a limited genetic base and that further improvement of this crop may require the introduction of genetic variability at DNA level during breeding programs. Further, the DNA polymorphisms may be or may not be associated with the morphological features of the somaclone shoots. Thus, in the current research RAPD markers were used to detect genetic diversity in C. peruvianus somaclones and to examine relationships among the somaclones and plants grown from seeds in cultivated populations, and a possible association of DNA fragments with somaclones' morphological traits. The RAPD technique was selected since demands little cost and time, and isolation and characterization of microsatellite loci is yet restrict to any cactus species.
MATERIAL AND METHODS
Shoots of forty-eight C. peruvianus somaclones at the Experimental Botanic Garden of State University of Maringá (Maringá PR Brazil; altitude 554.9m; 23º 25'S; 51º 25' W) and nine C. peruvianus cultivated plants (grown from seeds) at the Medicinal Plants Garden of the State University of Maringá (distance 1 000m) were used as samples for DNA extraction. All 12-year-old plants of C. peruvianus, grown from seeds (cultivated plants C1-C9), presented areoles in typically linear ribs. Thirty somaclones (S1, S2, S3, S5, S6, S7, S8, S9, S11, S18, S23, S24, S27, S28, S29, S30, S31, S41, S44, S46, S48, S49, S57, S60, S61, S64, S65, S67, S68, and S71) presented the typical shoots of species; 11 somaclones (S22, S26, S32, S35, S40, S45, S54, S55, S62, S66, and S69) presented atypical shoots, and 7 somaclones (S19, S33, S43, S47, S51, S53, and S59) had mixed shoot phenotypes (typical and atypical parts).
Fresh shoot sections (100-200 mg) from somaclones and cultivated plants were individually prepared according to protocol by Aljanabi et al. (1999), with minor modifi cations (higher NaCl concentration). Shoot sections were ground to a fi ne powder in liquid nitrogen and homogenized in 300-600 μL buffer 200 mM Tris-HCl, pH 8.0, 50 mM EDTA, pH 8.0, 5 M NaCl, 2% w/v CTAB, 0.06% sodium sulfi te, 20% w/v CTAB, 5% w/v lauril-sarcosine, and 10% w/v PVP-40. DNA was extracted with 1 volume of phenol: chloroform:isoamyl alcohol (25:24:1), incubated for 2h with 0.1 ng/μL of RNAse (10 ng/mL) and extracted with 1 volume of chloroform:isoamyl 5 M NaCl (0.06 volumes) were used for DNA precipitation. Isolated DNA quality was determined by electrophoresis in 0.8% agar gel (Hoisington et al., 1984). UV quantifi cation by visual comparison with known quantities of lambda DNA (Invitrogen) averaged about 20 ng/μL - 150 ng/ μL for each sample.
The amplification reactions were undertaken according to Williams et al. (1990), with minor modifi cations; they were performed in aseptic chambers using volumes of 20 μL containing 25 ng of genomic DNA, 10 mM Tris-HCl, pH 8.8, 2.5 mM MgCl2, 91 mM each of dATP, dGTP, dCTP, dTTP, 0.2 μL primer and 1 unit of Taq polymerase (Invitrogen). After amplifi cation reactions with OPA-12 and OPB-06 ten-mer primers (Operon Technologies Inc. Alameda CA USA), a total of 61 primers of kits OPA, OPB, OPC, OPF, OPL, OPM, and OPP were tested for amplifi cation reactions. Amplifications were performed, in duplicate, with personal Eppendorf Mastercycler Gradient. PCR conditions were: denaturation for 5 min at 96°C, 45 cycles of 94°C for 45 sec, 35°C for 1 min, and 72°C for 1 min and 30 sec, with a fi nal 7 min extension of 72°C.
Amplification products were separated by electrophoresis in 1.7% agar (Invitrogen) TAE gels at 60 V for 4 hr. Gels were stained with ethidium bromide (0.5 mg/mL) and registered by image captured in a High Performance Ultraviolet Transilluminator - Edas 290, using Kodak 1D 3.5 program. 1 Kb DNA Ladder (Invitrogen) was used as size marker.
Among the 61 primers tested, 16 (OPA-02, OPA-04, OPA-09, OPA-13, OPA-20, OPB-01, OPB-07, OPC-06, OPF-09, OPL-11, OPM-02, OPM-07, OPM-10, OPP-02, OPP-07, and OPP- 09) were used to amplify the DNA segments after an initial primer screening using DNA of the two somaclones and cultivated plants. The 16 primers, which yielded repetitive patterns for all scored bands, were applied to all 9 plants from cultivated population (C1, C2, C3, C4, C5, C6, C7, C8, C9) and to all 48 somaclones plants. Only one mix was prepared for each primer, which was used to compare simultaneously the 57 individual DNA samples for the same amplification reaction.
Fragments were analyzed by comparing RAPD profi les of each plant in terms of presence or absence of each DNA fragment. Plants' similarity was calculated by Jaccard's coeffi cient, while UPGMA cluster analysis was performed with NTSYS-pc software (Rohlf, 1989). The cophenetic correlation between the data of the genetic similarity matrix and cophenetic values of the dendrogram was estimated to indicate the dendrogram representativity in relation to original similarity estimated. All analyses were performed with NTSYS-pc software (Rohlf, 1989).
RESULTS
Polymorphic banding patterns in the Figure 1 illustrate the genomic variability in the somaclone plants (Table 1). Figure 1 illustrate the RAPD markers in somaclones observed using different primers (parts of the gels, from different amplifications reactions used to simultaneous comparison of the 57 individual samples).


Figure 1. RAPD fi ngerprinting of somaclones (S1 - S69) of C. peruvianus obtained with the primers OPM-02 and OPP-09 from Operon Technologies Inc. (Alameda, CA) on the samples of genomic DNA. The 1 kb ladder was used as molecular weight marker.
Table 1. Polymorphic Fragments in Somaclones and Cultivated Plants of Cereus peruvianus.

Where only the DNA segments most stronger stained were scored. The 16 primers generated 249 reproducible fragments (Table 1) of which 155 were polymorphic in the somaclones and 106 were polymorphic in the cultivated plants. The number of bands for each primer varied from 7 to 21, with an average of 15.6 fragments per primer. The size of amplifi ed products ranged from 300 to 5.500 bp. The OPA-04, OPB-07, and OPM-02 primers generated the highest number of fragments among the tested primers while primer OPB-07 showed the greatest capacity for discriminated polymorphic fragments (Table 1).
The dendrogram by Jaccard coeffi cient (Figure 2) showed that there were two grouping of somaclones at the time that cultivated plants were separately grouping; somaclones and cultivated plants were grouping according to their differential origins (in vitro regenerated and seed-grown C. peruvianus plants). Matrices of cophenetic values generated from RAPD data showed signifi cant and high correlation between the dendrograms obtained (r = 0.885) using the MxComp procedure from NTSYS program, which indicates a good fit between the original similarity matrix and the resulting clustering analysis. There was no grouping of somaclones according to typical or atypical shoot morphologies. Somaclones S22, S26, S32, S35, S40, S45, S54, S55, S62, S66, S69 with atypical shoots and S19, S33, S43, S47, S51, S53, S59 with mixed shoot phenotypes were casually grouping with the somaclones which present typical shoot morphologies. Casual grouping of somaclones with atypical and typical shoot phenotypes showed that the random amplifi ed DNA-fragment polymorphisms detected in present study were not associated with the shoot morphologies. Whereas OPB-07 primer produced a 300 pb fragment specifi cally detected in cultivated plants, the 750 pb fragment produced by OPP-09 was specifically detected in somaclone; the 1330 pb fragment produced by OPA-04 was specifi cally detected in a somaclone with mixed shoot phenotype (arrow in Figure 1A). However, no fragment was typical or atypical shoot-specific.
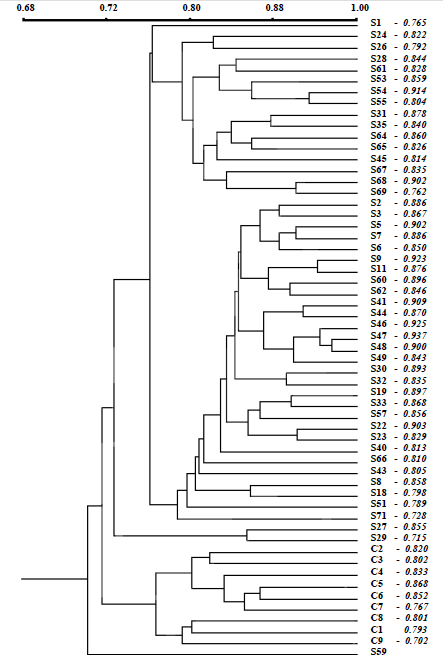
Figure 2. Dendrogram represents the relationship between the cultivated plants (C1 - C9) and the 48 somaclones (S) of C. peruvianus based on UPGMA cluster analysis of the RAPD profiles derived from 16 primers, by Jaccard's similarity coefficient.
Dendrogram also revealed that similarity in somaclones and cultivated plants ranged from 70.2 to 93.7% and 76.7 to 86.8%, respectively.
DISCUSSION
Polymorphism in somaclone plants (62.5%) was higher than that detected in cultivated plants (42.5%) and also higher than the polymorphism reported for descendents R1 of S3, S9, S48, and S71 somaclones (Resende et al., 2007) and for 633 regenerated plants of C. peruvianus using biochemical markers of 22 isozyme loci, which was 13.6% (Mangolin et al., 1997). The level of polymorphism in somaclones and cultivated plants was also higher than RAPD-detected polymorphism among 12 clones of C. peruvianus (11%) from California and 13 seedlings of C. jamacaru (9.4%) from northeastern Brazil (Gutman et al., 2001). Low level of genetic diversity found in species plants has been reported to be the result from vegetative propagation as the predominant form for multiplication of C. peruvianus populations (Nerd et al., 2002). Cultivation of the seed-grown C. peruvianus plants analyzed in current study is also practiced and mainly involves artificial selection in which desirable phenotypes are obtained by vegetative propagation. However, the primers selected in present study generated the highest number of fragments and showed a greater capacity for discriminated polymorphic fragments. This determined a higher level of DNA polymorphism in cultivated plants than DNA polymorphism in clones, reported by Gutman et al. (2001). In other columnar cactus species (Stenocereus stellatus, e.g., also used in central Mexico for its edible fruits), the cultivated populations in homegardens are more diverse than populations under silvicultural management (Casas et al., 2006). Thus, higher diversity might be expected in the C. peruvianus homegarden populations compared with that in populations managed in situ. Esterase polymorphism in F1 descendents from 14 cultivated populations has also shown high proportion of polymorphic loci (42.85%) and large genetic basis (from 0.4812 to 0.9657) for C. peruvianus (Faria et al. 2011).
In our research the reported higher DNA polymorphism between somaclones may be the result of induced genetic variability in vitro since somaclones were independently regenerated from different callus tissues. No association between RAPD markers and morphological variations has been detected in the somaclones of other plant species (Goto et al., 1998; Chen et al., 1998). Differential phenotypes in in vitro regenerated plants have been explained by different causes (Taji et al., 2002), e.g., differential methylation events. According to Kaeppler and Phillips (1993), differential methylation is a consequence of in vitro stress conditions. Hypomethylation has been associated with different morphological phenotypes in clonal regenerants of Solanum tuberosus (Joyce and Cassels, 2002) and Elaeis guineensis (Jaligot et al., 2002; Kubis et al., 2003). The phenotype reversion detected in C. peruvianus somaclones showing mixed morphologies was also described in somaclones from other plant species and was considered product of methylation events (Joyce et al., 2003).
In vitro stress conditions were also related with phenotype alterations in plants which were in vitro regenerated by Yu et al. (1999) and Tregear et al. (2002). Genomic stress caused by cultural conditions has been described as the major cause of transposable element activation (Chahal and Gosal, 2002). In fact, transposable elements are known to cause phenotype changes in plants and their activation during in vitro culture induces somaclonal variation. Hypoacethylation of histones determining differential genes expression in Arabidopsis has also determined phenotype alterations (Finnegan, 2001).
New varieties have been developed through somaclonal variations in tomato, sugarcane, celery, Brassica and sorghum (Karp, 1995). The morphological variations (37.5%) generated in shoots of somaclones conferred for the 10-yearold somaclones morphological traits of other three varieties and/or species of the genus Cereus: i) shoots in which the areoles are found in broken ribs forming alternate knobby regular space is a common trait of Cereus spp. knobby monstrose form; ii) shoots in which the areoles are found in broken ribs forming alternate knobby irregular space is a common trait of C. peruvianus monstrose form or C. monstruosus species; iii) shoots with areoles in tortuous or spiraled ribs is a common trait of C. peruvianus var. tortuosus. Thus, the simple and cost-effective tissue culture technique possesses greater potential for improvement of vegetatively propagated crops such as the C. peruvianus species. In vegetatively propagated plants the non-heritable phenotypes effects found in primary regenerants (R0) are important since they may be asexually propagated.
The results of our study are particularly important because shown that in vitro tissue culture of C. peruvianus may be recommended to broaden the species's genetic base. Additionally, there is a promising usefulness of somaclones in cross-pollination with cultivated plants since the analysis of fl oral biology and microsporogenesis in somaclones showed that the fl oral and reproductive characteristics and the somaclones' meiotic characteristics are similar to the fl oral, reproductive, and meiotic characteristics of the C. peruvianus natural populations (Ruvolo- Takasusuki et al., 2006; Silva et al., 2006). As self-incompatibility has been reported for C. peruvianus (Silva and Sazima, 1995), the cross-pollination system may be advantageous on somaclones and seed-grown C. peruvianus plants to determine the high level of the species's genetic polymorphism. Higher level of DNA polymorphism was detected in R1 descendents of somaclones (55.85%; Resende et al., 2007) than that in DNA polymorphism detected in cultivated plants (42.57%) used in current study. Therefore, a signifi cant part of the in vitro induced changes in somaclones should be sexually transferred to their seed-derived descendents.
In current study RAPD markers were not associated with the morphological features of the somaclone shoots. On the other hand, the genetic variability found in somaclones provides additional support for the occurrence of new alleles at the same locus (Mangolin et al., 1997). It is also important as a source for new traits that may be necessary as breeding programs proceed and/ or to conservation of species genetic resources; RAPD markers should be utilized to evaluate germplasm resources, helping breeders to determine the level of molecular similarity among plants in a cross, and also help plant collectors in programs of the specie-biodiversity conservation.
ACKNOWLEDGEMENTS
The authors would like to thank CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brasília DF Brazil) for its fi nancial support.
REFERENCES
1. Aljanabi, S.M., Forget, L., Dookun, A. (1999). An improved and rapid protocol for the isolation of polysaccharide and polyphenol-free sugarcane DNA. Plant Molecular Biology Reporter 17, 1-8. [ Links ]
2. Alvarez, M., Costa, S.C., Huber, A., Baron, M., Fontana, J.D. (1995). The cuticle of the cactus Cereus peruvianus as a source of a homo- -D-galacturonan. Applied Biochemistry and Biotechnology 51/52, 367-377. [ Links ]
3. Alvarez, M., Costa, S.C., Utumi, H., Huber, A., Beck, R., Fontana, J.D. (1992). The anionic glycan from the cactus Cereus peruvianus - structural features and potential uses. Applied Biochemistry and Biotechnology 34, 283- 295. [ Links ]
4. Barros, M.J., Nozaki, J. (2002). Pollutants abatement from effluents of paper and pulp industries by flocculation/coagulation and photochemical degradation. Quimica Nova 25, 736-740. [ Links ]
5. Casas, A., Cruse-Sanders, J., Morales, E., Otero- Arnaiz, A., Valiente-Banuet, A. (2006). Maintenance of phenotypic and genotypic diversity in managed populations of Stenocereus stellatus (Cactaceae) by indigenous peoples in Central México. Biodiversity and Conservation 15: 879-898. [ Links ]
6. Casas, A., Valiente-Banuet, A., Rojas-Martínez, A., Davila, P. (1999) Reproductive biology and the process of domestication of the columnar cactus Stenocereus stellatus in Central Mexico. American Journal of Botany 86, 534-542. [ Links ]
7. Chahal, G.S., Gosal, S.S. (2002). Tissue culture in crop development. In: Chahal, G.S. and Gosal, S.S. (eds.) Principles and Procedures of Plant Breeding. Pangbourne, UK: Alpha Science International Ltd pp 429-456. [ Links ]
8. Chen, W.H., Chen, T.M., Fu, Y.M., Hsieh, R.M., Chen, W.S. (1998). Studies on somaclonal variation in Phalaenopsis. Plant Cell Report 18, 7-13. [ Links ]
9. Faria-Sala, J., Mangolin, C.A., Franzoni, J., Machado, M.F.P.S. (2011). Esterase polymorphism and the analysis of genetic diversity and structure in cactus populations descended from Cereus peruvianus plants regenerated in vitro. Biochemical Genetics 49, online first (DOI 10.1007/s10528-010-9405-5). [ Links ]
10. Finnegan, E.J. (2001). Epialleles - a source of random variation in times of stress. Current Opinion Plant Biology 5, 101-106. [ Links ]
11. Gamborg, O.L., Miller, R.A., Ojima, K. (1968). Nutrient requirements of suspension cultures of soybean root cell. Experimental Cell Research 50, 151-158. [ Links ]
12. Goto, S., Thakur, R.C., Ishii, K. (1998). Determination of genetic stability in long-term micropropagated shoots of Pinus thunbergii Parl. using RAPD markers. Plant Cell Reports 18, 193-197. [ Links ]
13. Gutman, F., Bar-Zvi, D., Nerd, A., Mizrahi, Y. (2001). Molecular typing of Cereus peruvianus clones and their genetic relationships with other Cereus species evaluated by RAPD analysis. Journal Horticultural Science and Biotechnology 76, 709-713. [ Links ]
14. Hoisington, D., Khairallah, M., González-de- León, D. (1984). Laboratory Protocols: CIMMYT Applied Molecular Genetics Laboratory. Second Edition, Mexico, D.F., CIMMYT, 50 p. [ Links ]
15. Jain, S.M. (2001). Tissue culture-derived variation in crop improvement. Euphytica 118, 153-166. [ Links ]
16. Joyce, S.M., Cassels, A.C. (2002). Variation in potato microplant morphology in vitro and DNA methylation. Plant Cell, Tissue and Organ Culture 70, 125-137. [ Links ]
17. Joyce, S.M., Cassels, A.C., Jain, S.M. (2003). Stress and aberrant phenotypes in in vitro culture. Plant Cell, Tissue and Organ Culture 74, 103-121. [ Links ]
18. Kaeppler, S.M., Phillips, R.L. (1993). Tissue culture-induced DNA methylation variation in maize. PNAS 90, 8773-8776. [ Links ]
19. Karp, A. (1995). Somaclonal variation as a tool for crop improvement. Euphytica 85, 295- 302. [ Links ]
20. Larkin, P.J., Scowcroft, W.R. (1981). Somaclonal variation - a novel source of variability from cell culture for plant improvement. Theorical Applied Genetics 60, 197-214. [ Links ]
21. Machado, M.F.P.S., Mangolin, C.A., Oliveira- Collet, S.A. (2000). Somatic crossing-over can induce isozyme variation in somaclones of Cereus peruvianus Mill. (Cactaceae). Haseltonia 7, 77-80. [ Links ]
22. Mangolin, C.A., Prioli, A.J., Machado, M.F.P.S. (1997). Isozyme variability in plants regenerated from calli of Cereus peruvianus (Cactaceae). Biochemical Genetics 35, 189-204. [ Links ]
23. Murashige, T., Skoog, F.A. (1962). A revised medium for rapid growth and bio-essays with tobacco tissue culture. Physiology Plantarum 15, 474-479. [ Links ]
24. Nerd, A., Tel-Zur, N., Mizrahi, Y. (2002). Fruits of vine and columnar cacti. In: Nobel, P.S. (ed.) Cacti Biology and Uses. University of California Press Ltd, London, England, pp.185-197. [ Links ]
25. Nozaki, J., Messerschmidt, I., Rodrigues, D.G. (1993). Tannery waters cleaning with natural polyeletrolytes: chemical speciation studies of chromium. Archives of Biology and Technology 36, 761-770. [ Links ]
26. Oliveira, A.J.B., Machado, M.F.P.S. (2003). Alkaloid production by callus tissue cultures of Cereus peruvianus (Cactaceae). Applied Biochemistry and Biotechnology 104, 149- 155. [ Links ]
27. Oliveira, S.A., Machado, M.F.P.S., Prioli, A.J., Mangolin, C.A. (1995). In vitro propagation of Cereus peruvianus Mill. (Cactaceae). In Vitro Cell Developmental Biololgy Plant 31, 47-50. [ Links ]
28. Resende, A.G., Mangolin, C.A., Machado, M.F.P.S. (2007). Genetic diversity in F1 descendents of Cereus peruvianus Mill. (Cactaceae) somaclones regenerated in South Region of Brazil. Tropical and Subtropical Agroecosystems 7, 193-199. [ Links ]
29. Rohlf, F.J. (1989). NTSYS-pc Numerical taxonomy and multivariate analysis system, version 1.50 - 1989. New York Exeter Publ. New York. [ Links ]
30. Ruvolo-Takasuski, M.C.C., Mangolin, C.A., Machado, M.F.P.S. (2006). Somaclones of C. peruvianus Mill. (Cactaceae) may contribute towards the broadening of the species genetic basis. Resource Journal Botany 1, 19-23. [ Links ]
31. Silva, N., Machado, M.F.P.S., Mangolin, C.A., Pagliarini, M.S. (2006). Microsporogenesis in somaclones of Cereus peruvianus Mill. (Cactaceae). Journal of Plant Science 1, 8-13. [ Links ]
32. Silva, W.R., Sazima, M. (1995). Hawkmoth pollination in Cereus peruvianus, a columnar cactus from southeastern Brazil. Flora 190, 339-343. [ Links ]
33. Taji, A., Kumar, P., Lakshmanan, P. (2002). The origin, nature, and signifi cance of variation in tissue culture. In: Taji, A. (ed.) In Vitro Plant Breeding. Food Products Press, An Imprint of the Haworth Press Inc, Oxford, pp101-109. [ Links ]
34. Tregear, J.W., Morcillo, F., Richard, F., Berger, A., Singh, R., Cheah, S.C., Hartmann, C., Rival, A., Duval, Y. (2002). Characterization of a defensin gene expressed in oil palm infl orescences: induction during tissue culture and possible association with epigenetic somaclonal variation events. Journal Experimental Botany 53, 1387-1396. [ Links ]
35. Weiss, J., Nerd, A., Mizrahi, Y. (1993) Development of Cereus peruvianus (apple cactus) as a new crop for the Negev desert of Israel. In New crops (Janick, J. and Simon, J.E., eds.), Wiley: New York, pp. 471-486. [ Links ]
36. Weiss, J., Nerd, A., Mizrahi, Y. (1994). Flowering and pollination requirements in Cereus peruvianus cultivated in Israel. Israel Journal Plant Science 42, 149-158. [ Links ]
37. Williams, J.G.K., Kubelik, A.R., Livak, K.J., Rafalski, J.A., Tingey, S.V. (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acid Research 18, 6531-6535. [ Links ]
38. Yu, H.J., Moon, M.S., Lee, H.S. (1999). Analysis of cDNA expressed during fi rst cell division of petunia petal protoplast cultures using expressed sequences tags. Molecules and Cell 9, 258-264. [ Links ]














