Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
BAG. Journal of basic and applied genetics
On-line version ISSN 1852-6233
BAG, J. basic appl. genet. vol.21 no.1 Ciudad Autónoma de Buenos Aires Jan./June 2010
ARTÍCULOS ORIGINALES
Bovine μ-calpain (CAPN1) gene polymorphisms in Brangus and Brahman bulls
Soria LA1, Corva PM2, Huguet MJ1, Miño S3, Miquel, MC1
1Cátedra de Genética, Facultad de Ciencias Veterinarias, Universidad de Buenos Aires, Chorroarín 280. (1427) Ciudad Autónoma de Buenos Aires. Argentina
2Departamento de Producción Animal. Facultad de Ciencias Agrarias, Universidad Nacional de Mar del Plata. CC 276. (7620) Balcarce. Argentina
3Laboratorio de Virus Equinos. Instituto de Virología, Centro de Investigaciones en Ciencias Veterinarias y Agronómicas, INTA-Castelar. CC 25. (1712) Castelar. Argentina
Corresponding author: Liliana A. Soria Cátedra de Genética Facultad de Ciencias Veterinarias Universidad de Buenos Aires Chorroarín 280 (1427) Ciudad Autónoma de Buenos Aires. Argentina Phone/FAX: 54 (11) 4524-8474 lsoria@fvet.uba.ar
ABSTRACT
The bovine CAPN1 gene encodes the large subunit of μ-calpain, which is thought to be one of the most important enzymes involved in postmortem beef tenderization. Three SNPs in CAPN1 (SNP 316, SNP 530 and SNP 4751) have been associated with beef tenderness in different beef cattle breeds. The objective of this work was to implement genotyping strategies for CAPN1 markers as part of a project pursuing the identifi cation and validation of molecular markers associated with bovine meat quality and composition. Three PCR-RFLP methods were designed to determine genotypes of 64 bulls (11 Angus, 43 Brangus and 10 Brahman). Unexpected patterns resulting from the PCR-RFLP analysis at SNP 316 and SNP 530 were resolved by cloning and sequencing and lead to the discovery of three substitutions not previously described. These mutations could be useful in population studies, such as the determination of the relative contribution of the Angus and Brahman breeds to the Brangus.
Key Words: Beef cattle; Meat tenderness; U-calpain; Molecular markers; Single nucleotide polymorphism
RESUMEN
El gen CAPN1 en bovinos codifica la subunidad mayor de la μ-calpaina, la cual es una de las principales enzimas involucradas en la tiernización postmortem de la carne. Tres SNPs en el gen CAPN1 (SNP 316, SNP 530 y SNP 4751) se han asociado a la variabilidad en la terneza de la carne en diferentes razas bovinas. El objetivo de este trabajo fue desarrollar protocolos para la determinación de genotipos para los marcadores en CAPN1, como parte de un proyecto de identificación y validación de marcadores moleculares asociados con la composición y calidad de la carne. Tres métodos de PCR-RFLP se diseñaron para determinar los genotipos de 64 toros (11 Angus, 43 Brangus y 10 Brahman). La clonación y secuenciación de los productos de PCR correspondientes a toros con patrones de RFLP anómalos en los SNP 316 y SNP 530, revelaron tres sustituciones no descriptas previamente. Estas mutaciones podrían ser útiles para estudios poblacionales, como por ejemplo la determinación de la contribución relativa de Angus y Brahman en la raza Brangus.
INTRODUCTION
The μ-calpain is a cysteine protease that degrades myofibrillar proteins during postmortem storage of meat. The regulation of enzyme activity has been correlated with variation in meat tenderness (Geesink and Koohmaraie, 1999). The CAPN1 gene encodes the large subunit of this enzyme. Bovine CAPN1 has been mapped to chromosome 29 (Smith et al., 2000).
The SNP (Single Nucleotide Polymorphism) is a simple bi-allelic and co-dominant molecular marker that has been extensively used in association analyses with production traits in livestock (Vignal et al., 2002). Page et al. (2002) reported several SNPs by sequencing the 22 exons and 19 of the 21 introns of the bovine CAPN1 gene (GenBank Accession Nos. AF252504 and AF248054). The majority of the SNPs were found in introns or were synonymous substitutions, except one substitution in exon 9 (C/G) and another in exon 14 (G/A) (SNP 316 y SNP 530, respectively). The SNP 316 (alleles C/G) determines the replacement of Ala by Gly in the amino acid 316 of the protein (domain II) and the other (alleles G/A) causes the change of Ile by Val in the position 530 (domain III). These SNPs have been associated with differences in beef tenderness in a wide range of Bos taurus breeds (Page et al., 2004). In Bos indicus cattle both unfavorable alleles are close to fixation; however, other SNPs segregating in Brahman (Markers 4751, 4753 and 5331) have been identifi ed (Casas et al., 2005; White et al., 2005). The 4751 marker, a C/T substitution in intron 17, seems to be associated with tenderness in B. taurus, B. indicus, and their crosses (White et al., 2005). A recent study confirmed that SNP 316 and SNP 4751 had an effect on meat tenderness in different breeds (Van Eenennaam et al., 2007). Corva et al. (2007) confirmed the effect of SNP 316 and SNP 530 in B. taurus steers fattened on pastures in Argentina.
Brangus is a composite breed that was developed to combine favorable traits of Angus (B. taurus) and Brahman (B. indicus) cattle taking advantage of hybrid vigor at the same time. The Brahman breed is well known for its rusticity and maternal ability, whereas Angus is recognized for their superior carcass quality. While Brangus is appreciated for his resistance and adaptation, there are some concerns related to carcass quality, especially beef tenderness, and these problems become more important as the proportion of Bos indicus increases in the cross (Shackelford et al., 1995).
The aim of this work was to implement simple and inexpensive genotyping strategies for CAPN1 polymorphisms as part of a project of identification and validation of molecular markers associated with bovine meat quality and composition in beef cattle of Argentina. Our intention is to validate these and other SNPs as molecular markers of meat quality, especially for tenderness, in argentine Brangus cattle.
MATERIAL AND METHODS
Samples: Brangus (n=43), Angus (n=11) and Brahman (n=10) bulls used in Artifi cial Insemination (AI) were selected for this study. DNA was isolated from 400 μl of semen by standard methods using proteinase K and phenol/chloroform extraction and resuspended in Tris HCl 10 mM (Sambrook et al., 1989). The cell lysis buffer was modified as a 0.4 mM concentration of dithiothreitol.
Genotyping: Three PCR-RFLP methods were designed to determine genotypes at the SNPs 316, 530 and 4751. Primers were designed based on published CAPN1 sequences (GenBank Accession Nos. AF252504 and AF248054), using the Primer3 software (www.genome.wi.mit.edu). Fragments spanning each SNP were amplified by PCR. Primer sequences and PCR conditions appear in Table I. One hundred nanograms of DNA were submitted to 35 cycles of PCR with the following PCR program: 45 sec at 95°C, 45 sec at annealing temperature and 45 sec at 72°C, with a fi rst step at 95°C for 5 min and a fi nal extension for 5 min at 72°C. The PCR amplification was performed in a total volume of 25 μl containing 0.25 μM of each primer, 1-1.5 mM of MgCl2, 200μM of each dNTP (Promega Corporation, Madison, WI), IX reaction buffer and 1.25 U of proofreading DNA polymerase (InvitrogenTM Life technologies, Brazil). The PCR products were digested with an appropriate restriction enzyme (New England Biolabs, Beverly, MA) that discriminated alleles at each SNP. Two different restriction enzymes were used for SNP 530. Ambiguous results obtained with AvaII for heterozygotes were confirmed with PflFI (Table I). Twelve microliters of PCR product were digested with 2 units of restriction enzyme for 5 h and then electrophoresed in 1.8% agarose gels at 60V for 1 h 15 min.
Table I. PCR-RFLP tests for the genotyping of SNPs on the bovine CAPN1 gene.

DNA sequencing: The PCR products corresponding to SNP 316 and SNP 530 of four bulls that showed abnormal RFLP patterns (two for each SNP) were purifi ed with Qiaex® II Gel Extraction Kit (Qiagen, Hilden, Germany) and cloned in p-GEM®-T Easy Vector System II (Promega Corporation, Madison, WI). Clones were isolated with the Wizard® Plus SV Minipreps DNA Purification System (Promega Corporation, Madison, WI). Thirty positive clones were obtained. Plasmid DNA from thirteen positive clones was sequenced in an ABI3130XL Genetic Analyzer from both ends using T7 and SP6 primers.
Phylogenetic analyses: Two fragments of genomic DNA from CAPN1 located between positions 5251 to 5960 of sequence AF252504 (exons 8 to 10) and 4427 to 5206 of sequence AF248054 (exons 13 to 15) were analyzed separately. In order to perform a phylogenetic analysis, homologous bovine sequences were recovered from GenBank with the Basic Local Alignment Search Tool (BLAST: http://blast. ncbi.nlm.nih.gov, accessed in April 2010). Sequences shorter than 400 pb or with ambiguous sites were excluded. The sequences were assembled, edited and analyzed using the Bioedit software package version 7.0.9.0 (Hall et al., 1999). Sequences were aligned using Clustal X version 1.81 (Thompson et al., 1997) using default settings. Parsimony analyses were performed with the TNT software, version 1.1 (Goloboff et al., 2008). Two gap treatments were implemented. Gaps were considered missing data when they were located at the extremes of the alignment, but they were treated as a 5th state, when located in middle regions of the alignment, since they represented insertion-deletion events (Jones et al., 2004). The consistency of the identified groups was assessed by Bootstrap (Felsenstein, 1985) and Parsimony Jackknifing (Farris et al., 1996) tests.
RESULTS AND DISCUSSION
Sixty four bulls were genotyped for three different markers on the CAPN1 gene. In Brangus bulls, the allele frequency analysis showed a relatively moderate to high proportion of the allele favoring tenderness at each CAPN1 marker. Minor allele frequencies were 0.19 (C), 0.02 (A) and 0.45 (T) for SNP 316, SNP 530 and SNP 4571 respectively. A small proportion (4.7 %) had CC genotype at SNP 316. Genotype GG at SNP 530 was the most frequent (95.4%) and no homozygous AA bulls were found in the entire sample. The SNP 4751 showed three genotypes and the percentage of homozygous CC was 23.3% (Table II).
Table II. Genotypic frequencies (number of favorable alleles) and allele frequencies of three SNP related to beef tenderness.

Genotyping was performed according to the band pattern of digested PCR products (Table I). However, PCR-RFLP methods produced unexpected restriction patterns for some bulls, both for SNP 316 (enzyme BtgI) and SNP 530 (enzyme PflFI. Two Brangus bulls with anomalous RFLP for BtgI (Fig. 1) did not have the normal recognition site for this enzyme (ccatgg) located in intron 8. Two other bulls (one Brangus and one Brahman) showed unexpected RFLP patterns for PflFI in exon 14, while the recognition site of AvaII (ggwcc) remained unaffected (Fig. 3C). The PCR-RFLPs obtained with BtgI, AvaII and PflFI are shown in Fig.1 and Fig. 3. Initially, these abnormal results were attributed to partial digestions. However, cloning and sequencing confirmed that anomalous RFLPs patterns were generated by substitutions that abolished the BtgI and PflFI normal restriction sites. These mutations are the SNP 5340 (A/C) in intron 8 (base 5340 of AF252504) and the SNP 4554 (C/T) in exon 14 (base 4554 of AF248054) for BtgI and PflFI respectively. Thus the unexpected restriction patterns can be easily explained if the haplotypes defined at each region by the original tenderness markers and the novel SNPs are taken into account (Fig. 2 and Fig. 4).

Figure 1. Identification de SNP 316 alleles by PCR-RFLP using BtgI (A) and RFLPs of a Brangus bull with a mutation (SNP 5340) in the BtgI recognition site (B). A. Lane 1: DNA molecular weight marker (100bp Marker, Biodynamics); lanes 2, 3 and 4: bulls with different genotypes at SNP 316 that are all AA at SNP 5340; lane 5: bull with genotypes CA at SNP 5340 and GG at SNP 316; lane 6: negative control (PCR fragment without digestion). B. Lanes 2 and 3: clones with CG (Sequence DQ111767) and AG (Sequence DQ111768) haplotypes defi ned by SNP 5340 and SNP 316; lane 4: PCR product from genomic DNA with genotypes CA at SNP 5340 and GG at SNP 316; lane 5: negative control (PCR fragment without digestion).

Figure 2. Observed PCR-RFLP patterns using enzyme BtgI for the SNP 5340/SNP 316 haplotype in two Brangus bulls.

Figure 3. Identification de SNP 530 alleles by PCR-RFLP using AvaII (A) and Pfl FI (B) and RFLPs of a Brangus bull with a mutation in the PflFI recognition site (C). A and B: Lane 1: DNA molecular weight marker (100bp Marker, Biodynamics); lanes 2, 3 and 4: bulls with different genotypes at SNP 530 that are all CC at SNP 4554 and lane 5: negative control (PCR fragment without digestion). C. Lane 1: DNA molecular weight marker (100bp Marker, Biodynamics); lane 2: GG genotype at SNP 530, identifi ed with AvaII; lane 3: bull with genotypes CT at SNP 4554 and GG at SNP 530 (erroneously classifi ed as AG when PflFI is used). (Sequence DQ111769); lane 4: negative control (PCR fragment without digestion).

Figure 4. Observed PCR-RFLP patterns using enzyme AvaII for the SNP 530 and enzyme PfIFI for the SNP 4554/SNP 530 haplotype in two bulls (one Brangus and one Brahman).
Four sequences representing the novel haplotypes were selected from the available clones and submitted to GenBank (Accession Nos. DQ111767 to DQ111770). The identities of these cloned sequences were confirmed by independent forward and reverse sequencing of at least two clones, indicating that they represent genuine variation within the species, and are not the result of PCR or sequencing artifacts. These sequences shared high similarity with the published bovine CAPN1 sequences, confirming that they represent allelic variants of the bovine CAPN1 gene.
Sequencing of the regions spanning markers 316 and 530 revealed other mutations (substitutions and deletions) that distinguished them from the first published DNA sequences of CAPN1 gene (AF252504 and AF248054). These SNPs were found in different introns (Table III and IV). These mutations had been already reported in Bos taurus by other authors, with the exception of substitutions at positions 4731 (Table IV), 5340 and 5397 (Table III). The SNP 4554 that abolished the recognition site of PfIFI was identified when CAPN1 sequences were recovered from GenBank for a phylogenetic analysis, but not when the genotyping method for SNP 530 was designed. In that opportunity, PfIFI was chosen to discriminate between AA and AG genotypes in cases when it was not possible with AvaII.
Table III. Comparison between sequences DQ111767 and DQ111768 from two Brangus bulls, spanning exons 8 to 10 of bovine CAPN1 and consensus from 44 reported sequences.

Table IV. Comparison between sequences DQ111769 and DQ111770 from two bulls (Brangus and Brahman respectively), spanning exons 13 to 15 of bovine CAPN1 and consensus from 30 reported sequences.

The Brangus bulls that were analyzed in this study are extensively used by breeders. Therefore it is likely that the previously nor reported mutations are, or will soon become, actual polymorphisms segregating in the breed. At this point, we have information regarding SNP 5340 only. We found fi fteen individuals with the AC genotype in a sample of 246 Brangus steers from nine different commercial herds.
Although the novel SNPs are all located in introns, their functional significance should not be underestimated because they may influence mRNA stability and/or translation. Besides, these SNPs may be in linkage disequilibrium with polymorphisms in other regions of the CAPN1 gene with structural or functional significance.
The relationship of the sequences DQ111767 to DQ111770 with those recovered from GenBank was evaluated through a phylogenetic analysis (Fig. 5 and Fig. 6). The bootstrap analysis of the CAPN1 nucleic acid sequences corresponding to exons 8 to 10 identified four large and distinct branches (groups). Groups I to III correspond to allele G at SNP 316, whereas group IV corresponds to allele C. Detailed analysis of sequence heterogeneity revealed substantial nucleotide diversity at sites 5340, 5397, 5680, 5688, 5709, 5823 and 5902 (data not shown). The sequences DQ111767 and DQ111768 are in a different group, with haplotypes CACGGCA and AGCGGCA respectively (Fig. 5). These haplotypes were not found in the other sequences analyzed that originated from Bos taurus, suggesting that both correspond to Bos indicus breeds.
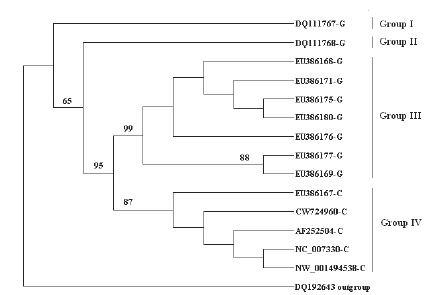
Figure 5. Phylogenetic tree showing the relationships among CAPN1 nucleic acid sequences corresponding to exon 8-exon 10. Numbers below key nodes indicate the percentage of bootstrap values of 100 replicates. Sequence DQ192643 (Sus scrofa CAPN1) was used as outgroup.

Figure 6. Phylogenetic tree showing the relationships among CAPN1 nucleic acid sequences corresponding to exon 13-exon 15. Numbers below key nodes indicate the percentage of bootstrap values of 100 replicates. All sequences analyzed have the G allele at SNP 530. Sequence DQ192544 (Sus scrofa CAPN1) was used as outgroup.
Figure 6 shows two distinct groups of sequences identified by the phylogenetic analysis of exons 13 to 15 of the CAPN1 gene. All the sequences of group I have a deletion of 4 nucleotides at positions 4733-4736. DQ111769 and DQ111770 have the haplotypes TGG----TA and TGA----CC, which are absent in the other sequences analyzed (from Bos taurus) and probably originated in Bos indicus.
Sequences from Bos indicus breeds are usually underrepresented in nucleotide databases. The results reported here suggest that to avoid misleading interpretations, special care must be taken for the development of genotyping strategies using sequences from Bos taurus breeds as a reference if they are supposed to be used in Bos indicus breeds an their crosses.
CONCLUSIONS
We identified novel substitutions on the bovine CAPN1 gene. These SNPs define haplotypes that probably originated in Bos indicus and could be considered in association studies for a highly important trait such as beef tenderness when working with composite breeds. The characterization of the novel mutations found in Brangus and Brahman bulls could also be useful in population genetics studies, such as the analysis of the relative contribution of Angus and Brahman to the Brangus breed.
REFERENCES
1. Casas E, White SN, Riley DG, Smith TP, Brenneman RA, Olson TA, Johnson DD, Coleman SW, Bennett GL, Chase CC Jr. (2005). Assessment of single nucleotide polymorphisms in genes residing on chromosomes 14 and 29 for association with carcass composition traits in Bos indicus cattle. J Anim Sci. 83 (1) :13-19. [ Links ]
2. Corva PM, Soria L, Villarreal EL, Schor A, Perez Cenci M, Motter M, Mezzadra C, Melucci LM, Paván E, Depetris G, Miquel MC, Santini FJ and Grigera Naón JJ. (2007). Association of polymorphisms on the CAPN1 and CAST genes with meat tenderness in beef cattle of Argentina. Genet. and Mol. Biol 30 (4) : 1064-1069. [ Links ]
3. Farris JS, Albert VA, Källersjö M, Lipscomb D, Kluge AG. (1996). Parsimony Jackknifing outperforms Neighbor-Joining. Cladistics, 12 : 99-124. [ Links ]
4. Felsenstein J. (1985). Confi dence limits on phylogenies, an approach using the Bootstrap. Evolution, 38 : 16-24. [ Links ]
5. Geesink GH, Koohmaraie M. (1999). Postmortem proteolysis and calpain/calpastatin activity in callipyge and normal lamb biceps femoris during extended postmortem storage. J Anim Sci. 77 : 1490-1501. [ Links ]
6. Goloboff P A, Farris J S, Nixon K C. (2008). TNT, a free program for phylogenetic analysis. Cladistics 24 : 774-786. Program and documentation, available from the authors, at www.zmuc.dk/public/phylogeni [ Links ]
7. Hall TA. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41 : 95-98. [ Links ]
8. Jones L R, Cigliano M M, Zandomeni RO, Weber EL. (2004). Phylogenetic analysis of bovine pestiviruses: testing the evolution of clinical symptoms. Cladistics 20 : 443-453. [ Links ]
9. Page BT, Casas E, Heaton MP, Cullen NG, Hyndman DL, Morris CA, Crawford AM, Wheeler TL, Koohmaraie M, Keele JW and TPL Smith. (2002). Evaluation of singlenucleotide polymorphisms in CAPN1 for association with meat tenderness en cattle. J. Anim. Sci. 80 (12) : 3077-3085. [ Links ]
10. Page BT, Casas E, Quaas RL, Thallman RM, Wheeler TL, Shackelford SD, Koohmaraie M, White SN, Bennett GL, Keele JW, Dikeman ME, Smith TP. (2004). Association of markers in the bovine CAPN1 gene with meat tenderness in large crossbred populations that sample infl uential industry sires. J Anim Sci. 82 (12) : 3474-3481. [ Links ]
11. Sambrook J, Fritsch, E F and Maniatis T. (1989). Molecular Cloning: a Laboratory Manual, 2nd ed. Appendix E3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [ Links ]
12. Shackelford SD, Wheeler TL, Koohmaraie M. (1995). Relationship between shear force and trained sensory panel tenderness ratings of 10 major muscles from Bos indicus and Bos taurus cattle. J Anim Sci. 73 (11) : 3333- 3340. [ Links ]
13. Smith TP, Casas E, Rexroad CE, Kappes SM, Keele JW. (2000). Bovine CAPN1 maps to a region of BTA29 containing a quantitative trait locus for meat tenderness. J Anim Sci. 78 (10) : 2589-2594. [ Links ]
14. Thompson JD, GibsonTJ, Plewniak F, Jeanmougin F, Higgins DG. (1997). The ClustalX windows interface: fl exible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res, 24 : 4876-4882. [ Links ]
15. Van Eenennaam A L, Li J, Thallman RM, Quaas R L, Dikeman M E, Gill C A, Franke D E and Thomas M (2007). Validation of commercial DNA test for quantitative beef quality traits. J Anim Sci. 85 (4) : 891-900. [ Links ]
16. Vignal A, Milan D, SanCristobal M, Eggen A. (2002). A review on SNP and other types of molecular markers and their use in animal genetics. Genet Sel Evol. 34 (3) : 275-305. [ Links ]
17. White SN, Casas E, Wheeler TL, Shackelford SD, Koohmaraie M, Riley DG, Chase CC Jr, Johnson DD, Keele JW, Smith TP. (2005). A new single nucleotide polymorphism in CAPN1 extends the current tenderness marker test to include cattle of Bos indicus, Bos taurus, and crossbred descent. J Anim Sci. 83 (9): 2001-2008. [ Links ]














