Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO
Related links
-
 Similars in
SciELO
Similars in
SciELO  uBio
uBio
Share
BAG. Journal of basic and applied genetics
On-line version ISSN 1852-6233
BAG, J. basic appl. genet. vol.23 no.2 Ciudad Autónoma de Buenos Aires Dec. 2012
RESEARCH
Taxonomy and phylogenetic analysis of Aspergillus section nigri isolated from yerba mate in Misiones (Argentina)
Castrillo M.L. 1*, Fonseca M.I. 1, Bich G.A. 1, Jerke G. 1, Horianski M.A. 1, Zapata, P.D. 1
1Laboratorio de Biotecnología Molecular. Módulo de Farmacia y Bioquímica. Facultad de Ciencias Exactas Químicas y Naturales, Universidad Nacional de Misiones.
Av. Mariano Moreno 1375. CP 3300. Posadas, Misiones, Argentina. Tel. +54 376 4427687.
* mlc_827@hotmail.com
ABSTRACT
Members of Aspergillus section Nigri are distributed worldwide and are potentially ochratoxin A producers; some of them are used at industrial level as source of extracellular enzymes and organic acids. The taxonomy of this section comprises one of the most complex and confusing of genus Aspergillus. The objectives of present work were to study the phylogeny of Aspergillus section Nigri and their taxonomy by amplifying ITS1, 5.8S and ITS2 regions. We analyzed 14 strains of Aspergillus section Nigri isolated from different commercial forms of yerba mate, classified according to morphology. Genomic DNA was extracted, the ITS regions were amplified with universal primers ITS1 and ITS4, and the obtained fragments were sequenced. The obtained sequences were aligned and edited to a size of 532bp, to carry out phylogenetic analyses using TNT program. We used 100 RAS+TBR cycles to search for the most parsimonious tree, which was supported by booststrap and Jackknife with 1000 resampled matrices. From the analysis of parsimony, one hundred trees were obtained with 568 steps. Bootstrap and Jackknife analyses exhibited similar topology and minor differences in their supported values, showing that section Nigri is distributed in two general clusters: one composed of A. niger, A. carbonarius and A. heteromorphus clades, and the other composed of A. aculeatus and A. homomorphus clades, separated from different sections of genus Aspergillus.
Key words: Aspergillus section Nigri; Taxonomy; Ribosomal DNA.
RESUMEN
Las especies de Aspergillus sección Nigri están distribuidas mundialmente y son consideradas potenciales productores de ocratoxina A; algunas de ellas se usan a nivel industrial como fuentes de enzimas extracelulares y ácidos orgánicos. La taxonomía de esta sección comprende una de las más complejas y confusas del género Aspergillus. El objetivo del presente trabajo fue estudiar la filogenia de Aspergillus sección Nigri y su taxonomía mediante la amplificación de la región ITS1, 5,8S e ITS2. Analizamos 14 cepas de Aspergillus sección Nigri asiladas de diferentes formas comerciales de yerba mate, clasificadas de acuerdo a su morfología. Se extrajo ADN genómico, las regiones ITS fueron amplificadas con los cebadores universales ITS1 e ITS4, y los fragmentos obtenidos fueron secuenciados. Las secuencias obtenidas fueron alineadas y editadas a un tamaño de 532pb, para realizar un análisis filogenético con el programa TNT. Para averiguar el árbol más parsimonioso, usamos 100 ciclos RAS+TBR, respaldados con 1000 repeticiones por análisis Bootstrap y Jackknife. Del análisis de parsimonia, se obtuvieron 100 árboles con 568 pasos. Ambos análisis, Bootstrap y Jackknife, mostraron topología similar y diferencias mínimas en sus valores respaldados, mostrando que la sección Nigri está distribuida en dos grupos generales: uno compuesto por los clados A. niger, A. carbonarius y A. heteromorphus; y el otro compuesto por los clados A. aculeatus y A. homomorphus, separados de las demás secciones del género Aspergillus.
Palabras clave: Aspergillus sección Nigri; Taxonomía; ADN ribosomal.
INTRODUCTION
The three major genera of mycotoxin-producing fungi are Aspergillus, Fusarium and Penicillium. These fungi frequently contaminate foods, usually causing severe biological alterations to human and animal health (Franco, 2005; Castrillo, 2010).
Members of Aspergillus section Nigri are the most important in the genus. They are ubiquitously distributed worldwide, growing on a wide variety of substrates and being considered as responsible fungi for food spoilage (Domsch et al., 1980; Pitt and Hocking, 1997; Abarca et al., 2004; Geiser et al., 2007). Some of them are a common source of extracellular enzymes and organic acids that are used in food processing (Yokotsuka and Sasaki, 1998; Abarca et al., 2004). Moreover, Aspergillus niger products hold the GRAS (Generally Recognized As Safe) status from the FDA (Federal Drugs and Foods Administration, USA) (Abarca et al., 2004). However, the potential capacity to produce mycotoxins by these species suppose a risk for human and animal health. Due to their biotechnological importance and because they are responsible of food spoilage, the identification of such strains needs to be unambiguous (Abarca et al., 2004).
The taxonomy of black aspergilli constitutes one of the most complex and confusing in genus Aspergillus; therefore, several taxonomic schemes have been proposed. Classical taxonomy for classification of species is primarily based on morphological criteria, such as forms and conidial ornamentations, size of conidia, conidiophore structures, and coloration of colonies, among others (Klich, 2002). Molecular techniques have contributed to these studies and allowed significant advances in fungal taxonomic organization. Ribosomal DNA (rDNA) amplification was one of the first applications of the Polymerase Chain Reaction (PCR) in mycology (White et al., 1990). The rDNA unit is constituted of regions that possess highly preserved genes, as the 18S and 28S regions, and universal primers were designed to allow internal transcribed spacers amplification of any fungus, independently of genus. The amplified region comprises ITS1, 5.8S and ITS2 regions, which can have variable nucleotide sequences (Fungaro, 2000), and are commonly used to examine phylogenetic positions or relationships at species or intraspecies level (Lee et al., 2000).
For many years, several authors have discussed the composition of section Nigri, taking into account morphological data and using rDNA sequences for phylogenetic analyses. Raper and Fennell (1965) produced the most comprehensive monograph of the genus and all Aspergillus species with conidial heads in some shades of black were included in the same group. They accepted twelve species and two varieties for Aspergillus section Nigri. Al-Musallam (1980) revised the taxonomy of this group of fungi using cluster analysis involving all available morphological and cultural parameters, and established five easily distinguishable species and the A. niger clade. The apparently insignificant differences between the members of A. niger clade was the decisive reason why Al-Musallam suggested that some of the species recognized by Raper and Fennell (1965) should be reclassified as varieties. Later, Kozakiewicz (1989) made a new proposal based upon conidial ornamentations, suggested alterations in the A. niger clade, and distinguished 16 taxa.
The previously mentioned classifications of Aspergillus section Nigri were solely based on morphological criteria and the delimitation of some taxa is problematic because they are distinguished by relatively small differences in variable characters (Parenicova et al., 2000; Varga et al., 2003; Abarca et al., 2004; Samson et al., 2004).
New molecular approaches have shown that there is a high biodiversity in Aspergillus section Nigri, but that species are occasionally difficult to recognize based solely on their phenotypic characters (Samson et al., 2007). Using this type of approaches, 19 species of Aspergillus section Nigri were accepted (Samson et al., 2007; Noonim et al., 2008; Perrone et al., 2008). More recently, Varga et al. (2011) reported that Aspergillus section Nigri includes 26 taxa.
Changes in the species concept of black aspergilli according to different authors are shown in Table 1.
As a further contribution, the aims of this paper were to study the phylogeny of Aspergillus section Nigri and their taxonomy by amplifying ITS1, 5.8S and ITS2 regions.
MATERIALS AND METHODS
Biological material
The strains used in this study were 14 isolates belonging to Aspergillus section Nigri of three commercial yerba mate forms: five strains from milled yerba mate, without ageing (referred as YMCHA); four strains from elaborated yerba mate (referred as YMA); and 5 strains from composed yerba mate (referred as YMCA). In addition, two Aspergillus carbonarius OTA+ standard strains with known capacity to produce ochratoxin were included, which were provided by the University of Buenos Aires (UBA).
The section Nigri strains initially were classified by microbiological classic methods, using Klich (2002) codes, by taking into account main macro- and micro-morphology criteria.
Extraction of genomic DNA
The fungal strains isolated from different commercial yerba mate forms were sown in YES (Yeast Extract Sucrose) liquid medium in order to obtain large amounts of mycelium. Mycelia was filtered and washed with Tris-HCl 0.1 M (pH 8) EDTA 0.02 M (pH 8). DNA extraction was carried out with buffer solution (Tris-HCl 0.1 M pH8, NaCl 1.5 M, EDTA 0.05 M pH8) at 60º C, containing Proteinase K 0.1mg/mL, ß-mercaptoethanol 10mM and SDS 2 % (p/v). DNA was purified with chloroform: isoamilyc alcohol (24:1 v/v) and potassium acetate 3M, and then was precipitated with isopropyl alcohol (Fonseca, 2012).
Amplification of ribosomal DNA
Ribosomal DNA fragments (ITS1 - 5.8S - ITS2) of 14 isolates of section Nigri and two A. carbonarius OTA+ standard strains, were amplified and sequenced. For the amplification reaction primers ITS1, 5' TTCGTAGGTGAACCTGCGG, and ITS4, 5' TCCTCCGCTTATTGATATGC (White et al., 1990) were used. The amplification reactions were prepared in a final volume of 20 µL, containing buffer 1X; MgCl2 2.5 mM; dNTP 200 µM; 10 pMol of each primer; Taq DNA polymerase 0.5 U and template DNA 5 ng/µL. PCR cycling was programmed to 35 cycles after an initial denaturation of 4 min at 94º C. Each amplification cycle consisted of the following steps: denaturation (94º C, 40 s), hybridization (50º C, 40 s), and extension (72º C, 40 s). Finally, a final extension of 10 min at 72º C was realized.
Agarose gels at 1% (p/v) and 2% (p/v) were performed, containing 10 mg/mL of ethidium bromide, visualized with UV transiluminator, and finally photographed with digital camera. Gels containing 1% (p/v) agarose were used to visualize genomic DNA and those with 2 % (p/v) agarose were used to visualize PCR products.
The concentration of DNA samples was semi-quantified by comparison to the intensity of the marker.
Sequencing of the regions ITS1 - 5.8S - ITS2
Sequencing reactions of PCR products were performed by Macrogen sequencing services (Kumchun-ku, Seoul, Korea).
Bioinformatic study of the obtained sequences
The obtained sequences were compared with those deposited in Database of the National Center for Biotechnology Information (NCBI, www.ncbi.nlm.nih.gov), with BLAST Search tool. Sequenced ITS1-5.8S-ITS2 regions and the sequences of interest retrieved from NCBI database were aligned by using the CLUSTAL W sequence editor on-line version (Thompson et al., 1994). The alignments were trimmed, overhangs were removed and gaps were corrected, prior to phylogenetic calculations with BIOEDIT software (Sequence Alignment Editor), for obtaining sequences of the same size and also for comparing the same regions. The sequences were submitted to Database of the National Center for Biotechnology Information (accession numbers: JF318957, JF436881, JF436882, JF436883, JF436884, JF436885, JF436886, JF436887, JF436888, JF436889, JF436890, JF436891, JF436892, JF436893, JF436894, JF436895).
To allow an appropriate phylogenetic analyses, sequences of Aspergillus section Nigri that were cited in Abarca et al. (2004) and Varga et al. (2011) were included in the present study (accession numbers: AJ280009, EU159211, AM087614, AJ280010, DQ900605, FJ491684, DQ900602, AJ280014, EU482439, FJ491679, FJ491683, AJ280013, EF166063, AY656625, JX291167.1, JQ316520.1, AJ279985, DQ900604, FJ491682, AJ223852, DQ900603, EU159216, HM853552, DQ900606, AJ223853, AY585549, FJ491678, L76747.1, AB000535, AF128852, AF203800, AJ000933, AY373938).
The parsimony analyses were performed with TNT version 1.1 (Willi Hennig Society Edition). Two methods were used: Bootstrap (Felsenstien, 1985) and Parsimony Jackknifing (Farris et al., 1996). Both analyses included 1000 resampled matrices. The existence of local optima for data sets beyond 40-50 taxa has long been well-informed (Maddison, 1991). In all analyses, Emericella nidulans CBS 12135 (AJ000933) was used as an out-group.
RESULTS
Analysis of genomic DNA amplification
Nucleic acid extraction of 14 fungal isolates from different commercial yerba mate forms (previously identified as species of Nigri section) and two A. carbonarius OTA+ standard strains were analyzed.
The PCR reaction with primers ITS1 and ITS4 amplified a fragment of approximately 600 bp for each isolate. The size of the amplified product was in close agreement with the expected fragment size in Aspergillus species (563 to 613 bp, according to White et al., 1990).
Bioinformatics study of the obtained sequences
The obtained sequences of each isolated were compared with deposited sequences in NCBI database, using BLAST as a search tool. In most cases, the high identity values obtained allowed us to assure that these species belong to Aspergillus section Nigri, always with identity values above 98 %. Seven strains of yerba mate (referred as YMCHA 73, YMCHA 71, YMCHA 55, YMA 120, YMA 119, YMA 10 and YMCA 29) were morphologically and molecularly defined as A. niger; three strains (referred as YMCHA 69, YMCA 18 and YMCA 2) were defined as A. tubingensis; and one strain (referred as YMCHA 63) was defined as A. brasiliensis; all strains belong to the A. niger clade. In addition, two standard strains (referred as PATRON I and PATRON II) were defined as A. carbonarius, that belongs to the A. carbonarius clade (Figure 1). However, three isolates of yerba mate (referred as YMCA 11, YMA 2 and YMCA 9) were morphologically defined only as Aspergillus section Nigri species, and molecularly defined as Aspergillus spp. (Figure 1).
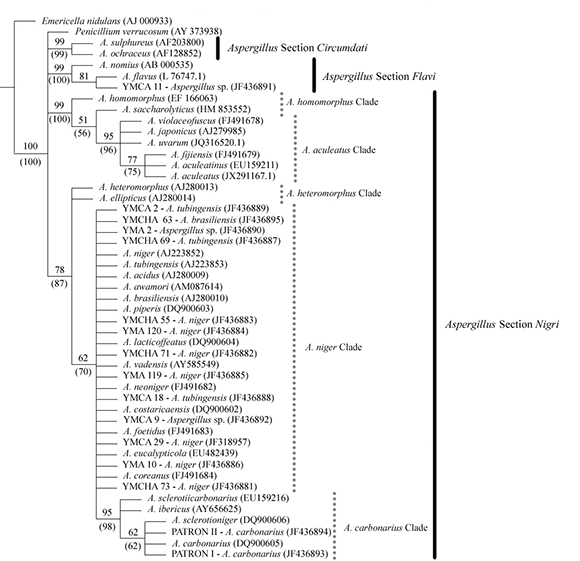
Figure 1. Phylogenetic analysis. Group support, assessed with 1000 Bootstrapping and Parsimony Jackknifing replicates.
Numbers above branches correspond to Bootstrap support. Jackknife supports are given in parentheses.
Phylogenetic Analyses
Good quality sequences of approximately 600 bp were first aligned and then trimmed, and sequences 532 bp in size were obtained, prior to carrying out phylogenetic calculations.
Study of parsimony was done using Bootstrap and Jackknife analyses. Parsimony analysis resulted in 100 equally parsimonious trees 568 steps long, performing 100 RAS + TBR cycles. Our data set consisted of 134 parsimony informative characters. Both, analyses, Bootstrap and Jackknife, yielded nearly the same topology, and presented minimal differences in support values. Our trees showed that all isolated strains of yerba mate are included in the A. niger and A. carbonarius clades, except for the isolated strain referred as YMCA 11 (Figure 1).
DISCUSSION
Until a few years ago, systematic studies of microorganisms were based almost exclusively on morphological criteria (classical microbiological classification). However, the number of tools actually used in systematic studies has increased. Nowadays, both morphological and molecular criteria are taken into account (Perez Valencia et al., 2005).
In this study, and for taxonomic organization of fungi, were considered the classical microbiological classification by morphological criteria and molecular techniques by amplification of ITS1, 5.8S and ITS2 regions. These regions are commonly used in fungi because they are composed of variable nucleotides sequences that allow the examination of phylogenetic positions or relationships at species or intraspecies level (Fungaro, 2000; Lee et al., 2000).
In Aspergillus section Nigri there are very subtle differences between species, being for this reason one of the most difficult groups for classification and identification. Consequently, classical microbiological criteria are insufficient for classification (Abarca et al., 2004; Varga et al., 2011). In this study, and by applying morphological criteria, we could accurately classify the isolated strains at the species level. However, it was necessary to confirm the morphological classification with molecular information obtained by amplification of ITS1, 5.8S and ITS2 regions and by contrasting them with those in NCBI database. Classical microbiological classification and molecular data ensured the correct strain classification at the genus level.
Most of the isolated strains belonged to Aspergillus section Nigri, invariably with high identity values, except in three cases (YMCA 11, YMA 2 y YMCA 9) which could only be morphologically defined as Aspergillus section Nigri and molecularly defined as Aspergillus spp., reflecting the confusing and complex taxonomy of this section. These exceptional cases either may belong to the A. niger aggregate or the bioinformatics analysis using solely the ITS conserved regions was insufficient for the correct molecular identification of Aspergillus section Nigri. Some authors have cited other conserved regions as useful for molecular classification, such as the calmodulin or parts of ß -tubulin sequences (Peterson, 2000; Abarca et al., 2004; Varga et al., 2011).
With respect to the phylogenetic analysis of Aspergillus section Nigri, in previous studies with amplification of ITS regions, Parenicova et al. (2000) reported that A. carbonarius could be easily distinguished from other black aspergilli. They cited that A. carbonarius forms a separate clade within the biseriate black aspergilli. Then, Parenicova et al. (2001) reported that ITS sequences could be used to distinguish uniseriate aspergilli in a separate clade from a general clade containing biseriate species and species belonging to section Flavi and Circumdati; but could not be used for differentiation within uniseriate species.
In this study, were observed four different general clades of Aspergillus strains, section Flavi, section Circumdati, and two general clusters of section Nigri: one general cluster containing A. aculeatus and A. homomorphus clades; and the other general cluster with A. heteromorphus clade and A. niger clade which includes A. carbonarius clade. A. carbonarius formed a separate branch into A. niger clade (biseriate species), as reported by Parenicova et al. (2000). But then, it was observed that in Aspergillus section Nigri, both biseriate (A. niger, A. carbonarius, A. heteromorphus and A. homomorphus clades) and uniseriate species (A. aculeatus clade) formed two general clades separated from section Flavi clade and section Circumdati clade (Figure 1). It was not observed that the uniseriate species formed a separate clade from all others, as previously cited by Parenicova et al. (2000).
Based on phylogenetic analysis of ITS sequence data, Varga et al. (2011) reported that Aspergillus section Nigri formed two general clusters: one cluster containing A. aculeatus and A. homomorphus clades and the other cluster containing A. heteromorphus clade and A. niger clade with A. carbonarius clade, as observed in this study.
In this phylogenetic analysis it is possible to visualize that some of the isolated strains of yerba mate belong to A. niger clade so it can be argued that YMA 2 and YMCA 9 strains, which were morphologically defined as Aspergillus section Nigri and molecularly defined as Aspergillus spp., also belong to section Nigri clade. Likewise, it was observed that YMCA 11 strain belongs to section Flavi; it was molecularly referred as A. flavus, being the only strain that was poorly characterized by classical microbiological methods (Figure 1).
In addition, parsimony analyses performed with Bootstrap and Jackknife methods, revealing minimal differences between the values obtained with both methods; this allows us to assure that the phylogenetic analyses performed were consistent and also they were in accordance with other current reports (Varga et al., 2011).
Moreover, in this study, and in order to observe whether toxin production had implications for phylogeny, were considered sequences of known species with OTA capability, as Penicillium verrucosum, Aspergillus ochraceus and patterns I and II strains. The results did not show that these species were part of the same clade, the ochratoxin A production does not seem to have implications on the phylogeny of ochratoxin A-producing species when ITS sequences were considered (Figure 1). Some authors reported that a chloroperoxidase gene presumably takes part in ochratoxin biosynthesis in P. verrucosum, so the use of this sequence on phylogenetic analysis can be useful for discriminate the ochratoxin A-producing species from others (Geiser et al., 2007; Varga et al., 2011).
Our results show that Aspergillus section Nigri has a polyphyletic origin and that it is composed of two general clusters, one containing A. aculeatus and A. homomorphus clades and the other containing A. heteromorphus clade and A. niger clade which includes A. carbonarius clade. Also, the uniseriate species (A. aculeatus clade) did not appear as a different group in relation to the biseriate species in Aspergillus genus phylogeny. We can also conclude that the taxonomic classification of Aspergillus section Nigri still remains as confusing and complex topic, and to date, it is necessary to take several precautions for studying the phylogeny of Aspergillus section Nigri.
LITERATURE CITED
1. Abarca M.L., Accensi F., Cano J., Cabañes F.J. (2004) Taxonomy and significance of black Aspergilli. Anton van Leeuwnhoek Inter. J. Gen. and Molec. Microbiol. 86: 33-49. [ Links ]
2. Al-Musalllam A. (1980) Revision of the black Aspergillus species. Thesis. University of Utrecht, The Netherlands. [ Links ]
3. Castrillo M.L. (2010) Caracterización morfológica y confirmación molecular de Aspergillus sección Nigri aislados de yerba mate (Ilex paraguariensis). Tesis de Grado. Fac. Cs. Exactas, Qcas. y Naturales, UNAM. Misiones, Argentina. [ Links ]
4. Domsch K.H., Gams W., Anderson T.H. (1980) Compendium of soil fungi. Acad. Press, London, UK. [ Links ]
5. Farris J.S., Albert V.A., Källersjö M., Lipscomb D., Kluge A.G. (1996) Parsimony Jackknifing outperforms Neighbor-Joining. Cladistics 12: 99-124. [ Links ]
6. Felsenstein J. (1985) Confidence limits on phylogenies, an approach using the Bootstrap. Evolution 38: 16-24. [ Links ]
7. Fonseca M.I. (2012) Utilización de hongos de pudrición blanca de la provincia de Misiones en procesos de biopulpado: Aspectos bioquímicos y moleculares de sistemas ligninolíticos involucrados y prospección biotecnológica. Tesis Doctoral, Fac. Bqca, Qca y Fcia, UNT. Tucumán, Argentina. [ Links ]
8. Franco E. (2005) Aspergillus sección Nigri, estudio fisiológico y molecular de especies ocratoxigénicas. Tesis Doctoral, Fac. Veterinaria, UAB. Barcelona, España. [ Links ]
9. Fungaro M.H.P. (2000) PCR na Micologia. Biotecnol. Ciênc. e Desenvol. 17: 12-16. [ Links ]
10. Geiser D.M., Klich M.A., Frisvad J.C., Peterson S.W., Varga J., Samsom R.A. (2007) The currente status of species recongnition and identification in Aspergillus. Study Mycol. 59: 1-10. [ Links ]
11. Klich M.A. (2002) Identification of common Aspergillus species. Centraalbureau voor schimmelcultures, Utrecht, The Netherlands. [ Links ]
12. Kozakiewicz Z. (1989) Aspergillus species on stored products. Mycological Papers 161: 1-188. [ Links ]
13. Lee J.S., Ko K.S., Jung H.S. (2000) Phylogenetic analysis of Xylaria based on nuclear ribosomal ITS1-5.8S-ITS2 sequences. FEMS Microbiology Letters 187: 89-93. [ Links ]
14. Maddison D. (1991) The discovery and importance of multiple islands of most parsimonious trees. Syst. Zool. 40: 315-328. [ Links ]
15. Noonim P., Mahakarnchanakul W., Varga J., Frisvad J.C., Samson R.A. (2008). Two new species of Aspergillus section Nigri from Thai coffee beans. International Journal of Systematic and Evolutionary Microbiology 58: 1727-1734. [ Links ]
16. Parenicova L., Kester H.C., Benen J.A., Visser J. (2000) Characterization of a novel endopolygalacturonase fron Aspergillus niger with unique kinetic properties. FEBS Lett. 2-3: 333-336. [ Links ]
17. Parenicova L., Skouboe P., Frisvad J., Samson R.A., Rossen L., ten Hoor-Suykerbuyk M., Visser J. (2001) Combined molecular and biochemical approach identifies Aspergillus japonicus and Aspergillus aculeatus as two species. Appl. Environ. Microbiol. 2: 521-527. [ Links ]
18. Pérez Valencia L.I., Santerre A., Villallobos Arámbula A.R., Galván Corona A., Torres-Torres M.G., Rodríguez Contreras A., Guzmán Dávalos L. (2005) Extracción de DNA y amplificación de secuencias del ITS del DNAr de Ganoderma (Fungí, Basidiomycetes) para su uso en el análisis filogenético. CUCBA. 8 pp. [ Links ]
19. Perrone G., Varga J., Susca A., Frisvad J.C., Stea G. (2008) Aspergillus uvarum sp. nov., an uniseriate black Aspergillus species isolated from grapes in Europe. International Journal of Systematic and Evolutionary Microbiology 58: 1032-1039. [ Links ]
20. Peterson S.T. (2000) Phylogenetic relationships in Aspergillus based on rDNA sequence analysis. In: Samson R.A., Pitt J.I. (Eds) Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Harw. Acad. Publishers, Amsterdam, The Netherlands, pp. 323-355. [ Links ]
21. Pitt J.I., Hocking A.D. (1997) Fungi and Food Spoilage. Ed. Blackie Acad. and Profess. London. UK. [ Links ]
22. Raper K.B., Fennell D.I. (1965) The genus Aspergillus. Willians and Wilkins, Baltimore. [ Links ]
23. Samson R.A., Houbraken J., Kuijpers A., Mick F.J., Frisvad J. (2004) New ochratoxin A or sclerotium species in Aspergillus section Nigri. Studies Mycol. 50: 46-61. [ Links ]
24. Samson R.A., Noonim P., Meijer M., Houbraken J., Frisvad J.C., Varga J. (2007) Diagnostic tools to identify black Aspergilli. Studies in Mycology 59: 129-146. [ Links ]
25. Thompson J.D., Higgins D.G., Gibson T.J. (1994) CLUSTAL W, improving the sensitivity of progresive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acids. Res. 22: 4673-4680. [ Links ]
26. Varga J., Rigó K., Tóth B., Téren J., Kozakiewicz Z. (2003) Evolutionary relationships among Aspergillus species producing economically important micotoxins. Food Technol. Biotechnol. 41: 29-36. [ Links ]
27. Varga J., Frisvad J.C., Kocsubé S., Brankovics B., Tóth B., Szigeti G., Samson R.A. (2011) New and revisited species in Aspergillus section Nigri. Studies Mycol. 69: 1-17. [ Links ]
28. White T.J., Burns T., Lee S., Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J. (Eds.) PCR Protocols: A guide to methods and applications. Acad Press, San Diego, pp. 315-322. [ Links ]
29. Yokotsuka T., Sasaki M. (1998) Fermented protein foods in the Orient: shoyu and miso in Japan. In: Wood B.J.B. (Ed.) Microbiology of fermented foods. Blackie Acad. and Profess, London. [ Links ]













