Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
BAG. Journal of basic and applied genetics
versión On-line ISSN 1852-6233
BAG, J. basic appl. genet. vol.28 no.2 Ciudad Autónoma de Buenos Aires dic. 2017
ARTÍCULOS ORIGINALES
Migrations, admixture and genetic diversity in Central Argentinian Patagonia: analysis of autosomal Alu polymorphisms
Migraciones, mestizaje y diversidad genética en la Patagonia Central Argentina: análisis de polimorfismos autosómicos Alu
Parolin M.L.1*, Zanetti D.2, Calò C.M.3, Esteban E.2, Avena S.4, Carnese F.R.4, Moral P.2
1 Instituto de Diversidad y Evolución Austral, CCT CENPAT-CONICET, Puerto Madryn, Argentina.
2 Departament Biologia Animal, Unitat d’Antropologia, Facultat de Biologia, Universitat de Barcelona, Spain.
3 Dipartimento di Scienze della vita e dell’ ambiente, Università de Cagliari, Sardegna Italia.
4 Universidad Maimónides e Instituto de Ciencias Antropológicas, Facultad de Filosofía y Letras, Universidad de Buenos Aires, Argentina.
* corresponding author: parolin@cenpat-conicet.gob.ar
Fecha de recepción: 20/02/2017
Fecha de aceptación de versión final: 25/10/2017
ABSTRACT
This study aimed to analyze autosomal Alu insertions in three localities from Patagonia Argentina belonging to the Andes region and the coast of the Chubut province. Knowledge of the genetic diversity of these populations, along with the genealogical data, will contribute to better understand historical information, differential migration process and bio-demographic composition of the Central Patagonia region. In order to achieve this objective, 16 autosomal Alu insertion polymorphisms were genotyped: ACE, APO-A1, TPA25, FXIIIB, A25, HS4.32, D1, HS4.69, HS2.43, Sb19.12, Yb8NBC120, Sb19.3, Yb8NBC125, Ya5NBC221, DM, and CD4. Our results showed that the Central Patagonia region presents a complex continental genetic admixture with marked Native American roots (39% ± 1.2), Eurasian (56% ± 1.73) and, to a lesser extent, African (5% ± 1.7). The genetic proximity of the Patagonian samples in relation to groups from Europe and Northern Africa, but with a displacement towards the native communities, constitutes a clear indicator of the differential admixture process that took place in different regions of Argentina. Moreover, genetic differences were observed between Patagonian localities and Bahía Blanca (Central region of Argentina). These observations warned us that population genetic constitution analysis cannot be approached without bearing in mind the regional particularities, which are the result of the different historical, migratory, social-economic and demographic processes that occurs in the country.
Key words: Alu insertion polymorphisms; Argentina Central Patagonia; Admixture; Migrations.
RESUMEN
Este estudio tiene como objetivo el análisis de las inserciones autosómicas Alu en tres localidades de la Patagonia argentina localizadas en la región andina y costera de la provincia de Chubut. El conocimiento de la diversidad genética de estas poblaciones, junto con los datos genealógicos, contribuirán a una mejor comprensión de la información histórica, los procesos migratorios diferenciales y la composición bio-demográfica de la región central Patagónica. Para alcanzar este objetivo se analizaron 16 polimorfismos autosómicos de inserción Alu: ACE, APO-A1, TPA25, FXIIIB, A25, HS4.32, D1, HS4.69, HS2.43, Sb19.12, Yb8NBC120, Sb19.3, Yb8NBC125, Ya5NBC221, DM y CD4. Nuestros resultados mostraron que la región central Patagónica presenta una mezcla genética continental compleja de marcadas raíces nativo americanas 39% (± 1.2), eurasiáticas 56% (± 1.73) y, en menor medida, africanas 5% (± 1.7). La proximidad genética de las muestras patagónicas a los grupos de Europa y del Norte de África, pero con un mayor desplazamiento hacia las comunidades nativas, constituye un claro indicador del proceso de mezcla diferencial que tuvo lugar en las distintas regiones de la Argentina. Por otra parte, las diferencias genéticas observadas entre las localidades de Patagonia y Bahía Blanca (región central de la Argentina), nos advierten que no puede analizarse la constitución genética de las poblaciones sin tener en cuenta las particularidades regionales, que son el resultado de los diferentes procesos históricos, migratorios, socio-económicos y demográficos que ocurrieron en el interior del país.
Palabras clave: Polimorfismos de inserción Alu; Patagonia Central Argentina; Mestizaje; Migraciones.
INTRODUCTION
Argentina has a history of widespread admixture among Native American, Eurasian and African groups, with particular characteristics in each region of the country (Toscanini et al., 2007; Avena et al., 2012; Parolin et al., 2013a, 2015a; García et al., 2015). Knowledge of the genetic composition of urban populations allows the reconstruction of the biological identity of their inhabitants, both at local and regional levels. At the same time, this information can be useful to understand the admixture events associated with different foundational histories, migrations and demographic changes that characterized the populations under study.
Previous studies carried out in the Central Patagonia region by means of blood systems analysis, showed an Eurasian contribution of 51%, a Native American contribution of 47%, and an African contribution of 2% in the Andean locality of Esquel (ESQ) (Avena et al., 2010). The corresponding ancestral contributions were 59%, 37% and 4% respectively in Comodoro Rivadavia (CR) (Avena et al., 2009) and 67%, 29% and 4% in Puerto Madryn (PM) (Parolin et al., 2013b). More recently, by means of the analysis of 99 ancestry informative autosomal markers (AIMs) in a combination of samples from ESQ and CR, the above genetic composition was estimated to be 52% Eurasian, 44% Native American, and 4% African (Avena et al., 2012). Likewise, through the study of 22 STR autosomal markers in samples from filiation cases from Chubut province, ancestral contributions were estimated as 50% European, 46% Native American and 4% African (Parolin et al., 2015b). Uniparental genetic data obtained in Central Patagonia (Avena et al., 2009, 2010; Parolin et al., 2013b, 2014), supports the idea that in Argentina the native maternal participation rises towards the North (83-94%) and South of the country, including Patagonia (53-78%), and decreases to an average of 45% in the Central region. Meanwhile, the native paternal contribution is observed at a mean of 6% in every urban population studied by our research team, except for Northwest Argentina (NWA), with 17.3% (Di Fabio Roca et al., 2016) and the Andean localities of ESQ, with 23% (Parolin et al., 2013a), and San Carlos de Bariloche (SCB), with 18% (Parolin et al., 2015a). Other authors have observed this same trend upon studying urban and rural areas from the center and North of Argentina (Martínez Marignac et al., 2004; Wang et al., 2008; García and Demarchi, 2009; Bailliet et al., 2011; Motti et al., 2013). There is also a strong correlation between genetic data and the donors´ genealogical information, and between genetic data and the historical and demographic data of each locality. In this regard, it has been observed that genetic contribution of foreign migrants (mainly from bordering countries) to the relatively small receptive populations, increases towards the South of the country. At local level, ESQ presents the largest proportion of grandparents and donors born in the same city and its area of influence, whereas TW and PM exhibit a larger number of internal migrants from other Argentinean regions, mainly from the center of the country, with largerEuropean ancestry (Parolin et al., 2013b). However, we point out the existence of an important Bolivian community inhabiting the lower valley of the Chubut River region (Rossini et al., 2007). In addition, TW, CR and SCB present a strong influence of migrations from bordering countries, particularly of Chilean origin, with strong Native American ancestry (Bandieri, 2001; INDEC: Censo Nacional de Población, Hogares y Viviendas 2010- http//indec.mecon.ar). Moreover, CR is the Patagonian city with the highest migratory flow from NWA, attracted by the large oil industry that characterizes the city. These differences observed from previous studies carried out in the interior of the same region and even in the same province, as it is the case of Chubut, cautioned us about the fact that the analysis of the genetic composition of the populations must take into consideration particularities of local and regional historical migrations.
Alu polymorphisms are repetitive DNA insertions of 300-1,000 bp which are spread along the human genome and constitute approximately 5% out of its total sequence. Their origin is estimated at around 65 million years BP and they are found exclusively in primates. These markers are especially useful in evolutionary studies because of a number of characteristics that they share: high stability, neutrality, known ancestral state and low propensity to exhibit homoplasy (Batzer and Deininger, 2002; Rishishwar et al., 2015). These properties make the analysis of Alu insertions a tool of great utility for the analysis of biological relations among populations, reconstruction of historical processes, and population structures. Moreover, they have been successfully used as ancestry markers to estimate the geographic descent of the human populations (Luizon et al., 2007).
Alu markers have been used to study Eurasian and African populations (Calò et al., 2005; González-Pérez et al., 2003, 2010; Hernando Herráez, 2010; Zanetti et al., 2014) and, in a lesser extent, Native American groups (Battilana et al., 2002, 2006; Dornelles et al., 2004; Mateus- Pereira et al., 2005; Gayà-Vidal et al., 2010; Gómez-Pérez et al., 2013). However, there are only three studies that make use of the Alu insertions as a tool for genetic-population analysis in Argentina: Martínez Marignac et al. (2004) analyzed two Alu markes and three autosomal SNPs to estimate continental admixture (European, African and native American ancestry) in a sample of La Plata city (Buenos Aires province), Resano et al. (2007) analyzed 19 Alu markers to estimate admixture fractions in a sample from Bahía Blanca city (Buenos Aires province), and Gómez-Pérez et al. (2011) analyzed eight autosomal Alu markers to characterize genetic diversity and population structure of the Jujuy province. Motivated by the demonstrated utility of the Alu insertions in population genetic studies, we analyzed 16 autosomal Alu insertion polymorphisms (ACE, APO-A1, TPA25, FXIIIB, A25, HS4.32, D1, HS4.69, HS2.43, Sb19.12, Yb8NBC120, Sb19.3, Yb8NBC125, Ya5NBC221, DM, and CD4) in three localities of Chubut province, Central Patagonia: one in the Andes region (Esquel) and the other two in the coast (Comodoro Rivadavia and Puerto Madryn). Knowledge of the genetic composition of these populations will contribute to understand differential migration and admixture processes in different region of Argentina. At the same time, the knowledge of the genetic structure of a particular population could be a useful tool in biomedical research and forensic sciences.
MATERIALS AND METHODS
The populations
DNA of 150 non-related individuals from the cities of Esquel (ESQ; N= 50), Comodoro Rivadavia (CR; N= 50) and Puerto Madryn (PM; N= 50) were analyzed. ESQ is situated in the Andes region, whereas the other two cities are in the coastal area of the Chubut province, Central Patagonia, Argentina (Figure 1). Blood samples were taken, with prior signed informed consent, from volunteer donors who attended the hemotherapy services at public and private hospitals in each locality and signed a prior informed consent. The participants were thoroughly informed of the scopes of the study and answered a questionnaire to obtain information about their birthplace and that of the two preceding generations (parents and grandparents). Five ml of whole blood were obtained from each donor and collected in vacutainer tubes with EDTA anticoagulant (Ethylene Diamine Tetra Acetic Acid). DNA was extracted by the phenol-chloroform method (Sambrook et al., 1989). The protocols and procedures employed in this study were reviewed and approved by bioethics and educational committees of the Regional Hospital of CR, Sub-zonal Hospital of ESQ, and Zonal Hospital of PM.

Figure 1. Geographical location of the studied populations.
Genotyping
Sixteen Alu insertion polymorphisms (FXIII, HS2.43, D1, HS4.69, A25, TPA25, APOA1, CD4, HS4.32, ACE, Sb19.3, Sb19.12, DM, Yb8NBC120, YbNBC125 and Ya5NBC221), distributed in nine autosomal chromosomes (Nº 1, 3, 6, 8, 11, 12, 17, 19, 22; Table 1), were genotyped. Genotypes were scored by PCR amplification and subsequent electrophoretic migration in agarose gels. Primer sequences and amplification conditions were as previously described (Batzer and Deininger, 1991; Stoneking et al., 1997; González-Pérez et al., 2010). Each PCR reaction included positive and negative amplification controls. Genotype assignments were made by direct observation of 2% agarose gels (CD4, DM and FXIIIB, at 1%) dyed with GelRed 0.5x under UV light.
Table 1. Amplification characteristics of Alu polymorphisms.

Ta: Annealing temperature; Chr: chromosome location; PCR product size of Alu-containing/Alu-lacking allele; pb: pairs of bases.
Statistical Analysis
Analysis of allelic frequencies, heterozygosity indexes, Hardy- Weinberg equilibrium, differences between population pairs (Fisher’s exact test), molecular analysis of variance (AMOVA) and the respective genetic distances (Reynolds et al., 1983) were performed by using the Genetix v.4.05 software (Belkhir et al., 2002) and Arlequin v3.1 (Excoffier and Lischer, 2010). Admixture estimations, based on the Eurasian-Native American-sub-Saharan trihybrid model, were performed using the ADMIX95 software (http://genetica.fmed.edu.uy/software.htm) which is based on the Chakraborty (1975) genetic identity method. Populations genetic affinities were analyzed by the construction of a principal components graph (PCA), based on Reynolds distances, by using the Rv3.1.1 FactoMineR (Josse, 2008) statistical package. The spatial genetic patterns were estimated through a geographically referenced spatial principal component analysis (sPCA). This method provides evidences of global or local genetic structures by considering Esquel (ESQ) 42°54’ S, 71°19’ O; Comodoro Rivadavia (CR): 45°52’ S, 67°30’ O, and Puerto Madryn (PM): 42°49’ S, 65°4’ O. The Patagonia region is shaded in gray. the geographic distances as well. Global patterns correspond to positive proper values, whereas negative proper values indicate local patterns that indicate considerable genetic differences among neighbors. The importance of global and local patterns was assessed by the Monte-Carlo test with 10,000 permutations, by using the R adegenet package (Jombart et al., 2008).
Comparisons with other world populations
In order to analyze the genetic structure of the studied populations and the inter-population relationship, the results were compared with those previously obtained in Bahía Blanca and in twelve worldwide populations by other authors (Figure 2). These analyses were carried out using data from 10 Alu insertion polymorphisms shared in the sixteen populations studied (ACE, HS4.69, APOA1, CD4, HS4.32, Sb19.3, Sb19.12, Yb8NBC120, YbNBC125 and Ya5NBC221).

Figure 2. Thirteen worldwide populations taken from the literature for comparative purposes. 1-BB: Bahía Blanca (Resano et al., 2007); 2-QUE: Quechua and 3-AYM: Aymara (Gayà-Vidal et al., 2010); 4-C.SP: Central Spain, 5-S.SP: Southern Spain, 6-BAL: Balearic Islands, 7-FRA: France, 8-SIC: Sicily, 9-GRE: Greece, 10-TUR: Turkey, 11-MOR: Morocco, 12-ALG: Algeria and 13-IC: Ivory Coast (González-Pérez et al., 2010).
RESULTS
The allelic frequencies of 16 Alu insertions estimated in three Central Patagonia populations are detailed in Table 2. The highest insertion frequencies were recorded for the APOA1 locus in CR (0.98) and for the Ya5NBC221 locus in ESQ (0.94). Towest frequencies were observed for the HS2.43 locus in the three studied populations (0.03 in CR to 0.07 in PM). Moreover, the lowest genetic diversity was recorded for the APOA1 locus in CR (H= 0.04) and the highest heterozygosity for the TPA25 locus (H= 0.62) in PM which, as expected, showed an intermediate value of insertion frequency (0.55). We performed 48 tests (16 loci analyzed/population; p-value=0.001).
Table 2. Alu insertion frequencies, heterozygosity and Hardy-Weinberg equilibrium in three localities of Central Patagonia (Exact test).

H: average heterozygosity; p-HW: Hardy-Weinberg equilibrium significance, Bonferroni correction (p-value 0.05/48= 0.001).
After Bonferroni correction, , we could not reject the Hardy-Weinberg equilibrium hypothesis for any of the sixteen Alu markers (nominal p>0.001). The AMOVA analysis indicated that most of the total genetic variation in the three studied populations was due to within population differences, whereas only 0.46% of the total genetic variation was due to differences among populations. The matrix of genetic distances based on Reynolds method (Reynolds et al., 1983) did not exhibit significant differences (FST p-values>0.05) among the studied Central Patagonia populations: CR vs. ESQ (FST= 0.0027; p-value= 0.56), CR vs. PM (FST= 0.0051; p-value 0.39), PM vs. ESQ (FST= 0.0102; p-value 0.32). The studied populations were compared with thirteen worldwide populations, arranged in five groups according to geographic-population criteria: Europe, Sub-Saharan Africa, North Africa, Admixed America and Native American (Figure 2). The hierarchic AMOVA analysis was significant (p-value<0.0001) with values of FST= 0.13 (difference among populations), FSC= 0.02 (differences among populations and within groups), and FCT= 0.11 (differences among population groups). These values might indicate the existence of a geographic structure in the analyzed populations. Such geographic structure was studied in more detail with an sPCA analysis. The values of the tests were, respectively, non significant for the global (p-value= 0.27), and significant for the local (p-value= 0.02). This differentiation found at local level might be due to the distinction between Northern Africa and sub- Saharan groups, since the test was not significant when the sample from the Ivory Coast (IC) was removed.
The genetic distance matrix obtained with Reynolds et al. (1983) index exhibited significant differences (p-fst<0.05) between the populations of the present study and those from the rest of the world. However, relatively low genetic distances were observed among the Central Patagonia and Bahía Blanca populations (variation range FST= 0.006-0.013), and between populations from Southern and Central Spain (variation range= 0.033- 0.048), whereas larger genetic distances were observed between the Patagonian and Native American populations (variation range= 0.094-0.122). On the other hand, the highest distances were observed between the Patagonian and Sub-Saharan (IC) populations (variation range= 0.192- 0.219), highlighting the outlier position of the latter. The graphic representation of the distances matrix first shows a cluster among the Central Patagonia samples and then with Southern Buenos Aires (BB), with an intermediate position among the Native American, Northern African and European groups (Figure 3A). The first two axes represented 73.4% of the total genetic variance. The first component, including 46.9% of the total variance, clustered the Argentinian populations in a position intermediate between the two Native Bolivian populations and the European and Northern African populations. The second component (26.4%) allowed the distinction of the sub- Saharan African population from the rest of the European, Northern African and South American populations. When the analysis was repeated to eliminate the effect of the Ivory Coast sample, the observed pattern was essentially the same (Figure 3B).

Figure 3A. Graphic representation of the genetic distances matrix based on Reynold et al. (1983) distances. Abbreviations of the analyzed populations are described in Figure 2.

Figure 3B. Graphic representation of the genetic distance matrix based on Reynold et al. (1983) distances excluding the South African sample (IC: Ivory Coast). Abbreviations of the analyzed populations are described in Figure 2.
Based on the trihybrid admixture model of the Latin American populations, the allelic frequencies of 10 Alu loci from Europeans (average C.SP, S.SP, and SIC), Africans (IC) and native Americans (average QUE and AYM) were used to represent the parental populations; the respective references are described in Figure 2. The total Central Patagonia sample had an Eurasian contribution of 56% (± 1.7%), African contribution of 5% (± 1.7%), and Native American contribution of 39% (± 1.2%). At the local level, those contributions were respectively 58% Eurasian (± 2.4%), 1.5% African (± 2.2%), and 40.5% Native American (± 2.0%) in CR city. In ESQ, the contributions were 54.4% (± 5.5%) Eurasian, 3.3% (± 1.2%) African and 42.3% (± 3.6%) Native. Finally, in PM, those values were 56% (± 1.2%) Eurasian, 5% (± 4.8%) African, and 39% (± 1.0%) Native American. Genealogical information obtained for each sample demonstrates local differences in the relationship between the place of birth and geographic origin of the grandparents. In this sense, Figure 4 shows that the origin of the grandparents is mainly local in ESQ, from the Central region in PM, and from Chile in CR, whereas the origin ranking in the second place is Chile in PM, and Europe in CR and PM. In a lesser extent, other South American grandparents came from Bolivia, Paraguay, Uruguay, and Brazil.

Figure 4. Birthplace of grandparents.
DISCUSSION
This research constitutes the first genetic characterization of urban populations in the Argentinian Patagonia by means of analysis of Alu insertion polymorphisms. The Argentininian population presents a widespread multiple continental admixture with Native American, Eurasian and, to a lesser extent, African roots. In this sense, the Central Patagonian region is not an exception, as it has been shown previously in studies of uniparental and biparental genetic heritage markers (Avena et al., 2009, 2010, 2012; Parolin et al., 2013a,b, 2014, 2015a,b). In order to complement the previously mentionedstudies, 16 Alu markers were analyzed. The insertion frequencies and the average heterozygosity values were similar to those previously recorded in the Argentinian locality of Bahía Blanca (Resano et al., 2007) and, in general, in Northern Africa and Spain populations (González-Perez et al., 2010). Nevertheless, the CD4 locus presented a higher insertion frequency and a significantly lower heterozygosity value than in previous studies. In this regard, it has been observed that the insertion frequency of CD4 is close to 1.00 in native groups from South America (Gayà-Vidal et al., 2010). The comparatively high frequency of the ancestral allele CD4 might be related to the autoctonous contribution in the analyzed populations. Regarding the genetic differentiation observed at local level, the variation observed in the analyzed Alu markers only presents local discontinuities in the African continent, while these markers are not indicators of significant genetic heterogeneities in the other continents.
The admixture estimation based on the trihybrid model showed that the Central Patagonian sample registered an autoctonous contribution (36.5% ± 1.9%) higher than the one previously obtained for Southern Buenos Aires province (20.9% ± 4.8%). This fact might be related to the late incorporation of Patagonia to the country (Law 1532, 1884), which allowed the native populations to keep their autonomy for a longer period than other native groups. It could also be related to the constant migratory flow that the Patagonian region receives from inland provinces and from bordering countries, mainly from Chile, whose populations has a large native ancestry (Bandieri, 2001; Eyheramendy et al., 2015; Gómez-Carballa et al., 2016). Nonetheless, possible variations due to chance related to sample size and to the type and quantity of the analyzed genetic markers cannot be discarded. Likewise, the admixture results obtained at the local level are related to the geographic origin of the ancestors and to the foundational history of each analyzed population. This is reflected in the genealogical data that show clear differences in the geographical origin of the migrant grandparents by locality. In this sense, it was observed that, in ESQ, there is a higher frequency of birth in which the grandparents were also locally born. This fact could be related to a slow growth of the region and a better preservation of the autoctonous origin (Finkelstein and Novella, 2006). Since before its foundation (1906) and to present days, ESQ has maintained an intense trade with Chile, being a zone of transport and gateway of migrants in both sides of the Andes mountain range. ESQ has had a gradual and diversified growth, also having agroforestry and tourism as its most important economic activities (Avena et al., 2010). CR has had significant contributions of grandparents from Chile and northern provinces of Argentina. Both immigrant groups, which have large native ancestry (Avena et al., 2012; Bandieri, 2001; Eyheramendy et al., 2015; Gómez-Carballa et al., 2016; Gómez-Pérez et al., 2011; Ramallo et al., 2011), participated in strong migratory waves attracted by the exponential growth of the oil industry, which started in 1907 and is still ongoing (Avena et al., 2009).
On the other hand, PM presents the higher proportion of grandparents from the central region of the country (30%), who havemostly European ancestry (Avena et al., 2012), and 23% grandparents born in that continent. This fact is related to the migration of qualified laborers for the aluminum industry, mainly from the inner provinces of the country, and also to the foundational history of PM, with Welsh, Spanish and Italian roots. Nonetheless, we must indicate that, during the last two decades, an important migration from Boliviahas taken place, representing the migrants from that country 36% of the total coming from Latin America (INDEC: www.indec.mecon.ar). The Bolivian community harbors a considerable fraction of autoctonous ancestry (Heinz et al., 2013) and, nowadays, it is estimated that they represent 5.6% of the South American migrants in the Patagonian region (Castillo and Gurrieri, 2012). The three cities included in then present study have less than 150 years of existence; therefore the migrations have been very important in the growth of their populations. ESQ is located in a much more humid area than the other two cities, at the foot of Andes mountain range. PM and CR are located inthe coast, in a much drier area. The density of the native population was much higher in the mountain range than in the coast; this also explains that the native contribution has a local origin, being high the correspondence with the recorded genealogical information. Thus, although the percentage of Native American contribution was similar in the three cities, the source of that contribution is different.
The genetic affinities among the populations based on the Reynolds et al. (1983) distances and their graphical representation in the correspondent PCA show a genetic proximity between the studied samples and the Bahía Blanca population. This may be due in part to the geographic nexus between Patagonia and Buenos Aires province, which favors a migratory flow between both regions. In a similar way, the genetic proximity of the Central Patagonian samples and the Northern African and European groups, but with a shift towards the autoctonous communities, constitutes a clear indicator of the character of the differential admixture process that took place in the different regions of Argentina. Even though no significant differences have been observed among the populations of the Central Patagonia region, we would like to highlight the importance of having genealogical data to better understand and contextualize the genetic data. This information together with the historical and demographic data caution us that the analysis of the genetic makeup of the populations should be approached bearing in mind the local and regional particularities, which are the result of different historical, migratory, and social andeconomic processes. For example, this information helps us to understand that the main source of the autoctonous contribution was local in ESQ, Chilean in CR and of various origins in PM.
Finally, our results highlight the usefulness of Alu markers as a tool for the study of the biological relations and the analysis of genetic population admixture. Our results are also in agreement with the Eurasian, African and Native American contributions previously observed in the same populations for blood group systems (Avena et al., 2009, 2010; Parolin et al., 2013b), or using STRs (Parolin et al., 2015b) and in 99 individual ancestry informative markers (Avena et al., 2012) (Figure 5). In conclusion, this study reveals the utility of Alu insertion polymorphisms as a complementary tool for population genetic analysis, being a simple and economical molecular technique. Also, this study shows the need to increase biparental genetic information for American urban and rural populations, which will contribute to better understand the complex migratory and heterogeneous admixture processes observed in Latin American populations.
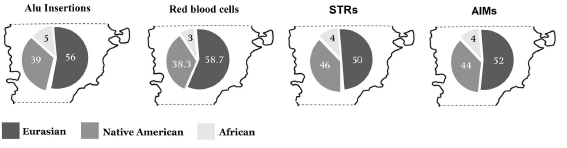
Figure 5. Percentage of the estimated genetic admixture in the Central Patagonia region for four autosomal systems.
BIBLIOGRAPHY
1. Avena S.A., Parolin M.L., Boquet M., Dejean C.B., Postillone M.B., Alvarez Trentini Y., Di Fabio Rocca F., Mansilla F., Jones L., Dugoujon J.M., Carnese F.R. (2010) Mezcla génica y linajes uniparentales en Esquel (Prov. de Chubut). Su comparación con otras muestras poblacionales Argentinas. J. Basic Appl. Genet. 21: 01-14. [ Links ]
2. Avena S.A., Parolin M.L., Dejean C.B., Ríos Part M.C., Fabrikant G., Goicoechea A.S., Dugoujon J.M., Carnese F.R. (2009) Mezcla génica y linajes uniparentales en Comodoro Rivadavia (Provincia de Chubut, Argentina). Rev. Arg. Antrop. Biol. 11: 25-41. [ Links ]
3. Avena S., Via M., Ziv E., Gignoux C.R., Dejean C., Huntsman S., Torres-Mejía G., Dutil J., Matta J.L., Beckman K., Burchard E.G., Parolin M.L., Goicoechea A., Acreche N., Boquet M., Fernandez V., Rey J., Stern M.C., Carnese R.F., Fejerman L. (2012) Heterogeneity in genetic admixture across different regions of Argentina. Plos One 7 (4): e34695. [ Links ]
4. Bailliet G., Ramallo V., Muzzio M., Santos M.R., Motti J.M.B., Bianchi N.O., Bravi C.M. (2011) Antecedentes y nuevos aportes en el estudio del Cromosoma Y en poblaciones humanas Sudamericanas. J. Basic Appl. Genet. 22: 1-9. [ Links ]
5. Bandieri S. (2001) Cruzando la cordillera: La frontera argentino-chilena como espacio social. En: Torres SB. (Ed.) La migración chilena en la Patagonia Austral en la primera mitad del siglo XX y su inserción en los centros urbanos de Comodoro Rivadavia, Río Gallegos y Ushuaia. Centro de Estudios de Historia Regional (CEHIR), Facultad de Humanidades, Universidad del Comahue, Neuquén, Argentina pp. 421-458. [ Links ]
6. Battilana J., Bonatto S.L., Freitas L.B., Hutz M.H., Weimer T.A., Callegari-Jacques S.M., Batzer M.A., Hill K., Hurtado A.M., Tsuneto L.T., Petzl-Erler M.L., Salzano F.M. (2002) Alu insertions versus blood group plus protein genetic variability in four Amerindian populations. Ann. Hum. Biol. 29: 334-347. [ Links ]
7. Battilana J., Fagundes N.J., Heller A.H., Goldani A., Freitas L.B., Tarazona-Santos E., Munkhbat B., Munkhtuvshin N., Krylov M., Benevolenskaia L., Arnett F.C., Batzer M.A., Deininger P.L., Salzano F.M., Bonatto S.L. (2006) Alu insertion polymorphisms in Native Americans and related Asian populations. Ann. Hum. Biol. 33: 142-60. [ Links ]
8. Batzer M.A., Deininger P.L. (1991) A human-specific subfamily of Alu sequences. Genomics 9: 481-487. [ Links ]
9. Batzer M.A., Deininger P.L. (2002) Alu repeats and human genomic diversity. Nat. Rev. Genet. 3: 370-379. [ Links ]
10. Belkhir K., Borsa P., Chikhi L., Raufaste N., Bonhomme F. (2002) GENETIX 4.03, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier, France. [ Links ]
11. Calò C.M., Piras I.S., Moral P., Falchi A., Ghiani M.E., Varesi L., Vona G. (2005) Analisi molecolare delle popolazioni del Mediterraneo attraverso 11 inserzioni Alu. Antrop. 9: 1-12. [ Links ]
12. Castillo J., Gurrieri J. (2012) El panorama de las migraciones limítrofes y del Perú en la Argentina en el inicio del siglo XXI. En: El impacto de las migraciones en Argentina. Cuadernos migratorios N° 2. Artola J., Gurrieri J., Ezequiel T., Camino A., Vaca V. (Eds.) Organización Internacional para las Migraciones. Editorial Oficina regional para América del Sur, Ciudad Autónoma de Jujuy pp. 365. [ Links ]
13. Chakraborty R. (1975) Estimation of race admixture: A new method. Am. J. Phys. Anthropol. 42: 507-511. [ Links ]
14. Di Fabio Roca D.F., Albeza M.V., Postillone B.M., Acreche N., Lafage L., Parolín M.L., Dejean C., Carnese F.R., Avena S. (2016) Historia poblacional y análisis antropogenético de la ciudad de Salta. ANDES: Antropología e Historia. En prensa. [ Links ]
15. Dornelles C.L., Battilana J., Fagundes N.J.R., Freitas L.B., Bonatto S.L., Salzano F.M. (2004) Mitochondrial DNA and Alu insertions in a genetically peculiar population: The Ayoreo Indians of Bolivia and Paraguay. Am. J. Hum. Biol. 16: 479-488. [ Links ]
16. Excoffier L., Lischer H.L. (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10: 564-7. [ Links ]
17. Eyheramendy S., Martinez F.M., Vial C., Repetto G.M. (2015) Genetic structure characterization of Chileans reflects historical immigration patterns. Nature Communications 6: 6472. [ Links ]
18. Finkelstein D., Novella M.M. (2006) Actividades económicas y proceso de construcción social en las áreas andinas de Río Negro y Chubut. En: Bandieri S., Blanco G., Varela G. (Eds.) Hecho en Patagonia. La historia en perspectiva regional. Editorial Educo- Universidad Nacional del Comahue, Neuquén, pp. 191-209. [ Links ]
19. García A., Demarchi D.A. (2009) Incidence and Distribution of Native American mtDNA haplogroups in Central Argentina. Hum. Biol. 81: 59-69. [ Links ]
20. García A., Dermarchi D., Tovo-Rodrigues L., Pauro M., Callegari-Jacques S., Salzano F., Hutz M. (2015) High interpopulation homogeneity in Central Argentina As Assessed by Ancestry Informative Markers (AIMs). Gen. Mol. Biol. 38: 324-331. [ Links ]
21. Gayà-Vidal M., Dugoujon L.M., Esteban E., Athanasiadis G., Rodríguez A., Villena M., Vasquez R., Moral P. (2010) Autosomal and X Chromosome Alu insertions in Bolivian Aymaras and Quechuas: two languages and one genetic pool. Hum. Biol. 22: 154-62. [ Links ]
22. Gómez-Carballa A., Moreno F., Álvarez-Iglesias V., Martinón-Torres F., García-Magariños M., Pantoja- Astudillo J.A., Aguirre-Morales E., Bustos P., Salas A. (2016) Revealing latitudinal patterns of mitochondrial DNA diversity in Chileans. Forensic Sci. Int.Genet. 20: 81-88. [ Links ]
23. Gómez-Pérez L., Alfonso-Sánchez M.A., Dipierri J.A., Alfaro E., García-Obregón S., De Pancorbo M.M., Bailliet G., Peña J.A. (2011) Microevolutionary processes due to landscape features in the province of Jujuy (Argentina). Am. J. Hum. Biol. 23: 177-84. [ Links ]
24. Gómez-Pérez L., Alfonso-Sánchez M.A., Dipierri, J.E., Sánchez D., Espinosa I., De Pancorbo M.M., Peña J.A. (2013) Young Alu insertions within the MHC class I region in Native American populations: Insights into the origin of the MHC-Alu repeats. Am. J. Hum. Biol. 25: 359-365. doi:10.1002/ajhb.2237. [ Links ]
25. González-Pérez E., Via M., Esteban E., López-Alomar A., Mazieres S., Harich N., Kandil M., Dugoujon J.M., Moral P. (2003) Alu insertions in the Iberian Peninsula and north west Africa-genetic boundaries or melting pot. Coll. Antropol. 27: 491-500. [ Links ]
26. González-Pérez E., Esteban E., Via M., Gayà-Vidal M., Athanasiadis G., Dugoujon J.M., Luna F., Mesa M.S., Fuster V., Kandil M., Harich N., Bissar-Tadmouri N., Saetta A., Moral P. (2010) Population relationships in the Mediterranean revealed by autosomal genetic data (Alu and Alu/STR compound systems). Phys. Anthropol. 141: 430-439. [ Links ]
27. Heinz T., Álvarez-Iglesias V., Pardo-Seco J., Taboada- Echalar P., Gómez-Carballa A., Torres-Balanza A., Rocabado O., Carracedo A., Vullo C., Salas A. (2013) Ancestry analysis reveals a predominant Native American component with moderate European admixture in Bolivians. Forensic Sci. Int. Genet. 7: 537-542. [ Links ]
28. Hernando Herráez I. (2010) Caracterización genética de las poblaciones de Valencia y Menorca mediante polimorfismos Alu. Tesis de Maestría en Biología Humana, Departamento de Biología Animal, Facultad de Biología, Universidad de Barcelona, España. [ Links ]
29. INDEC (2010) Censo Nacional de Población, Hogares y Viviendas. www.indec.mecon.ar. [ Links ]
30. Jombart T., Devillard S., Dufour A.B., Pontier D. (2008) Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity. Edinb. 101: 92-103. [ Links ]
31. Josse J. (2008) FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 25: 1-18. [ Links ]
32. Luizon M.R., Mendes-Junior C.T., De Oliveira S.F, Simões A.L. (2007) Ancestry informative markers in Amerindians from Brazilian Amazon. Am. J. Hum. Biol. Off. J. Hum. Biol. Counc. 20: 86-90. [ Links ]
33. Martínez Marignac V.L., Bertoni B., Parra E.J., Bianchi N.O. (2004) Characterization of admixture in an urban sample from Buenos Aires, Argentina, using uniparentally and biparentally inherited genetic markers. Hum. Biol. 76: 543-57. [ Links ]
34. Mateus Pereira L.H., Socorro A., Fernandez I., Masleh M., Vidal D., Bianchi N.O., Bonatto S.L., Salzano F.M., Herrera R.J. (2005) Phylogenetic information in polymorphic L1 and Alu insertions from East Asians and Native American populations. Am. J. Phys. Anthropol. 1: 171-184. [ Links ]
35. Motti J.M.B., Muzzio M., Rodenak Kladniew B., Alfaro E.L., Dipierri J.E., Bailliet G., Bravi C.M. (2013) Origen y distribución espacial de linajes maternos nativos en el noroeste y centro oeste argentinos. Rev. Arg. Antrop. Biol. 15: 3-14. [ Links ]
36. Parolin M.L., Avena S.A., Di Fabio Rocca F. (2013a) Análisis de la variación regional en el proceso de mestizaje de la Argentina. J. Basic Appl. Genet. Supp. 24: 30. [ Links ]
37. Parolin M.L., Avena S.A., Fleischer S., Pretell M., Di Fabio Rocca F., Rodríguez D.A., Dejean C.B., Postillone M.B., Vaccaro M.S., Dahinten S.L., ManeraG., Carnese F.R. (2013b) Análisis de la diversidad biológica y mestizaje en la ciudad de Puerto Madryn (Prov. de Chubut, Argentina). Rev. Arg. Antrop. Biol. 15: 61-75. [ Links ]
38. Parolin M.L., Basso N.G., Avena S.A. (2014) Mestizaje en la Patagonia Argentina: Diversidad genética y poblamiento de Trelew (Prov. Chubut). Basic Appl. Genet. Supp. 25: 250. [ Links ]
39. Parolin M.L., Toscanini U., Llull. C. (2015a) No tan aislados: el componente nativo presente en poblaciones cosmopolitas de la Patagonia Argentina. Simposio: Historia y prehistoria de los Pueblos Patagónicos contada por el ADN. Libro de resúmenes, XLVIII Reunión Anual de la Sociedad de Genética de Chile, Valdivia, e9. [ Links ]
40. Parolin M.L., Real L.E., Martinazzo L.B., Basso N.G. (2015b) Population genetic analyses of the Powerplex® Fusion kit in a cosmopolitan sample of Chubut Province (Patagonia Argentina). Forensic Sci. Int. Genet. 19: 221-222. [ Links ]
41. Ramallo V., Salceda S., Bailliet G. (2011) Apellidos, familias y linajes moleculares: el caso de Azampay, Catamarca. RUNA 23: 163-184. [ Links ]
42. Resano M., Esteban E., González-Pérez E., Vía M., Athanasiadis G., Avena S., Goicoechea A., Bartomioli M., Fernández V., Cabrera A., Dejean C., Carnese F., Moral P. (2007) How many populations set foot through the Patagonian door? Genetic composition of the current population of Bahía Blanca (Argentina) based on data from 19 Alu polymorphisms. Am. J. Hum. Biol. 19: 827-385. [ Links ]
43. R Core Team (2017) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.Rproject.org. [ Links ]
44. Reynolds J., Weir B.S., Cockerham C.C. (1983) Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics 105: 767-779. [ Links ]
45. Rishishwar L., Tellez Villa C.E., King Jordan I. (2015) Transposable element polymorphisms recapitulate human evolution. Mob DNA doi:10.1186/s13100- 015-0052-6. [ Links ]
46. Rossini O., Bonelli L., Kovacic G. (2007) Chagas en Puerto Madryn, Chubut. Retrospectiva de los últimos 12 años. http://chubut.gov.ar. [ Links ]
47. Sambrook J., Fritisch E.F., Maniatis T. (1989) Molecular Cloning: a laboratory Manual. 2nd Edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [ Links ]
48. Stoneking M., Fontius J.J., Clifford S.L., Soodyall H., Arcot S.S., Saha N., Jenkins T., Tahir M.A., Deininger P.L., Batzer M.A. (1997) Alu insertion polymorphisms and human evolution: evidence for a larger population size in Africa. Genome Res. 7: 1061-1071. [ Links ]
49. Toscanini U., Gusmão L., Berardi G., Amorim A., Carracedo A., Salas A., Raimondi E. (2007) Testing for genetic structure in different urban Argentinian populations. Forensic Sci. Int. 165: 35-40. [ Links ]
50. Wang S., Ray N., Rojas W., Parra M.V., Bedoya G., Gallo C. (2008) Geographic Patterns of Genome Admixture in Latin American Mestizos. PLoS Genet. 4: e1000037. doi:10.1371/journal.pgen.1000037. [ Links ]
51. Zanetti D., Sadiq M., Carreras-Torres R., Khabour O., Alkaraki A., Esteban E., Via M., Moral P. (2014) Human Diversity in Jordan: Polymorphic Alu Insertions in General Jordanian and Bedouin Groups. Hum. Biol. 86: 131-138. [ Links ]














