Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO  uBio
uBio
Compartir
BAG. Journal of basic and applied genetics
versión On-line ISSN 1852-6233
BAG, J. basic appl. genet. vol.29 no.2 Ciudad Autónoma de Buenos Aires dic. 2018
ARTÍCULOS ORIGINALES
Morphological and molecular characterization of Isostichopus sp. in the Colombian Caribbean sea
Caracterización morfológica y molecular de Isostichopus sp. en el mar Caribe Colombiano
Vergara, W.1, Agudelo, V.1, Castro, L.2, R. Rodríguez A.1*, Eeckhaut, I.3
1 Aquaculture Laboratory, Grupo de Investigación y Desarrollo Tecnológico en Acuicultura, Fisheries Engineering Program. Universidad del Magdalena. Cra. 32 No 22–08, Santa Marta, Colombia.
2 Grupo de Investigación en Evolución, Sistemática y Ecología Molecular. Biology Program. Universidad del Magdalena. Cra. 32 No. 22 - 08, Santa Marta, Magdalena.
3 Laboratoire de Biologie des organismes marins et Biomimétisme, Umons, 7000 Mons, Belgique.
* Corresponding author: arodriguezf@unimagdalena.edu.co
Fecha de recepción: 23/07/2018
Fecha de aceptación de versión final: 07/12/2018
ABSTRACT
Isostichopus sp. (Aspidochirotida: Stichopodidae) are sea cucumbers widely distributed in the Caribbean Sea. Amongst them, Isostichopus badionotus is one of the most harvested species. It shows a wide range of morphotypes widespread in the Caribbean region including different habitats (muddy, sandy and rocky bottoms, and sea grass beds). In Colombia, three morphotypes can be distinguished; two of them live in sea grass beds while the third one is found on rocky substrates. The present study describes the morphological characteristics of these morphotypes and analyzes their genetic structures through 16S rDNA and COI data. Our phylogenetic analyses show that the morphotype living on rocky substrates is morphologically and genetically distinct from the two other morphotypes and might not correspond to I. badionotus, the only species of the Isostichopus genus previously reported for this region.
Key words: Cucumbers; COI; 16S rDNA; Isostichopus badionotus.
RESUMEN
Isostichopus sp. (Aspidochirotida:Stichopodidae) son pepinos de mar ampliamente distribuidos en el mar Caribe. Entre ellos, Isostichopus badionotus es una de las especies mas cosechadas. Muestra un amplio rango de morfotipos extendidos en la region caribena, incluyendo diferentes habitats (fondos barrosos, arenosos y rocosos, y bancos de pastos marinos). En Colombia, se pueden distinguir tres morfotipos; dos de ellos viven en los bancos de pastos marinos mientras que el tercero se halla sobre sustratos rocosos. En el presente estudio se describen las caracteristicas morfologicas de esos morfotipos y se analizan sus estructuras geneticas mediante datos de 16S rDNA y COI. Nuestros analisis filogeneticos muestran que el morfotipo que vive sobre sustratos rocosos es morfologica y geneticamente distinto de los otros dos morfotipos y puede no corresponder a I. badionotus, la unica especie del genero Isostichopus previamente reportada para esta region.
Palabras clave: Pepinos; COI; 16S rDNA; Isostichopus badionotus.
INTRODUCTION
Sea cucumbers are over-exploited in the three main oceans, and started to be recently targeted in the Caribean sea (Purcell et al., 2012). Isostichopus badionotus is one of the sea cucumber species widespread in the Caribbean region (Guzman et al., 2003), ranging from North Carolina to northern Brazil. In addition, this species has been previously reported in the east to the middle-Atlantic and in the Gulf of Guinea (western Africa) (Hendler et al., 1995). It is found at depths from 0-55m (Miller and Pawson, 1990). I. badionotus is a common shallow water species that inhabits rocky bottoms, as well as sea grass beds (Hendler et al., 1995). In Colombia, there is an illegal, unregulated and unquantified fishery (Toral-Granda, 2008), and it is of potential commercial interest in Florida, Puerto Rico and the U.S. Virgin Islands (Bruckner, 2006).
At present, Colombian Caribbean Sea cucumbers have not been well studied (Rodriguez et al., 2013; Agudelo and Rodriguez, 2015) and there are many gaps in their knowledge especially related to their taxonomic characterization (Honey-Escandon et al., 2012; Smirnov, 2012). The advances in this regard allowed the reporting of around 44 species, most of which have been captured by exploratory fishing, and identified mainly as deep-sea species (Caycedo, 1978; Borrero et al., 2003; Borrero-Perez et al., 2012; Toral-Granda, 2008; Toral-Granda et al., 2008).
Since sea cucumbers are an important source of animal proteins, studies in Colombia are beginning to develop the conditions for their production under controlled environments (Rodriguez et al., 2013; Agudelo and Rodriguez, 2015; Vergara and Rodriguez, 2015; 2016). In this way, part of the present team studied the gonad morphology and the larval development of a species, Isostichopus sp. aff badionotus, including its spawning period (Agudelo-Martinez and Rodriguez-Forero, 2017). According to Guzman and Guevara (2002) and Toral- Granda (2008), there are three species of Isostichopus including I. badionotus, I. fuscus, and I. macroparentheses distributed in Central and South America. However, the observation of various morphotypes inside the species groups suggests the occurrence of more than three species for this region. In Colombia, three morphotypes can be distinguished in I. badionotus. The morphotypes studied are similar because they all have a robust body, a mouth located ventrally, twenty pelleted tentacles, anus in terminal position, gonads in the form of tufts, podias or ambulatory feet distributed in three rows, one or two poly vesicles, all these, distinctive characteristics of the Stichopodidae family. Preliminary phylogenetic analyses showed two morphotypes that cluster together and one that is well separated from the others. Two of them live in sea grass beds while the third is found on rocky substrates. Also, histological studies revealed a different structure of the body wall. The present study describes the morphological characteristics of these morphotypes and analyzes their genetic structures through 16S rDNA and COI data.
This research can be very useful to increase the knowledge about the taxonomy status of some Colombian sea cucumbers and to find out the morpho-anatomical and genetic differences between species of genus Isostichopus, from the native populations of the Santa Marta, Caribbean Sea. This information will also be useful for conservation and management purposes.
MATERIAL AND METHODS
Collection of animals
During one year, two hundred sea cucumbers were purchased from local artisanal fishermen in Rodadero Bay (11° 13′ 22.73″ N - 74° 13′ 32.59″ W), Airport Bay (11° 07′ 10″ N - 74° 13′ 50″ W), and Taganga Bay (11° 16′ 03.4″ N - 74° 11’ 32.3″ W), located in Santa Marta, Magdalena, Colombia (Figure 1) and transferred to the Aquaculture Laboratory (Universidad del Magdalena, Colombia). Most of the material was collected in shallow waters (1 to 20 m depth). One hundred and thirty sea cucumbers were collected between Rodadero and Airport bays while seventy were obtained from the bay of Taganga. Individuals from the three morphotypes were weighed with an analytical scale Ohaus (0.001 g), and their length was measured with a standard measuring board (mm). Photographs of the dorsal and ventral sides of the body were taken by using digital camera (Cannon EOS, Rebel XTi). Besides the external and internal anatomical observations, skin samples, tentacles and podia were taken for reviewing ossicles. Samples were dissolved in 10% sodium hypochlorite for 10 min and rinsed with water as described by Lambert (1985), and examined microscopically through slide preparations. For this, the animals were anesthetized by immersion in sea water with ice and then sacrificed. The characterization was made by comparison with Tommasi (1969) and Purcell et al., (2012). Our results showed that the specimens of the three morphotypes came from two lineages of Isostichopus species (Isostichopus badionotus and Isostichopus sp., characterized in this report) that were recorded in the Center of Biological Collections of Universidad de Magdalena (Santa Marta, Magdalena, Colombia) (CBUMAG:ECH:00001, CBUMAG:ECH:00002, CBUMAG:ECH:00003).
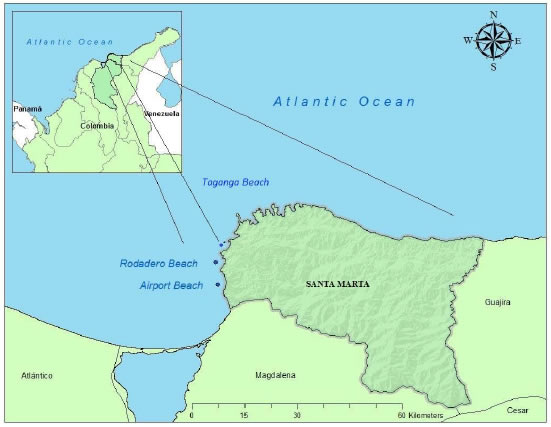
Figure 1. Marine and coastal areas of Colombia Caribbean Sea (modified by José Viillacob). Blue points in the studied areas.
DNA extraction, amplification and sequencing
Twenty-seven individuals were dissected and preserved in absolute ethanol (99.5%). DNA extraction was performed from up to 15 mg of muscle tissue using the DNeasy Blood & Tissue Kit (QIAGEN). The quality of the DNA was verified on a 1% agarose gel stained with GelRed (Biotium). PCR amplification of the COI gene was performed using the primers co1eF (5’-ATAATGATAGGAGGRTTTGG-3’) and co1eR (5’-GCTCGTGTRTCTACRTCCAT-3’) (Arndt et al., 1996), and of the 16S gene with the primers 16Sas (5’-CGCCTGTTTATCAAAAACAT-3’) and 16Sbr (5’-CTCCGGTTTGAACTCAGATCA-3’). PCR reactions were performed with 2 μL template in a 25 μL volume with final concentrations of 2 mM MgCl2, 5X PCR buffer (no MgCl2 BIOLINER), 0.4 μM of each primer, 0.4 μM of each dNTP, and 2 units Taq (BIOLASETM, BIOLINER). PCR amplifications were performed under the following conditions: 1 min at 95 °C, followed by 35 cycles of 15 s at 95 °C, 1 min at 40 °C, 1.5 min at 72 °C, and there was a final extension period of 5 min at 72 °C. The PCR products were verified on a 2% agarose gel stained with GelRed (Biotium). Following amplification, PCR products were purified using the MACHEREY-NAGEL kit (NucleoSpinR Extract II), and both strands were sequenced in both directions.
Sequence analyses and molecular phylogenetic analyses
Primer sequences were removed from the start and the end of the obtained sequence and sequence ambiguities were resolved by comparing the electropherograms using the program BioEdit v. 7.0.5.3 (Hall, 1999). After trimming, forward and reverse sequences for each specimen were assembled. Each assembled sequence was examined and edited by hand, and each sequence was checked for stop codons (in the case of the COI data file). Finally the consensus sequence from each contig was verified using the Blast tool on NCBI (www.ncbi.nlm.nih.gov). The obtained sequences and downloaded sequences from GenBank were aligned using the ClustalW algorithm (Thompson et al., 1997) in MEGA 6 (Tamura et al., 2013). The downloaded sequences were, for the COI data set, one sequence of I. badionotus (EU848276.1), one COI sequence of Isostichopus sp. (FJ971400.1), five sequences of I. fuscus (AF486424.1/28.1), three sequences of S. herrmanni, and one sequence of Holothuria leucospilota (KC405565.1); for the 16S data set, one sequence of I. macroparentesis (AY338415.1), two sequences of I. badionotus (JN207495.1, EU822435.1), six sequences of I. fuscus (AY153494.1, AY153499.1), three sequences of Stichopus herrmanni (EU822451.1, EU856636.1, FJ223863.1), and one sequence of Holothuria leucospilota (JQ657266.1). The COI final alignment was of 665 bp, and the 16S final alignment was of 508 bp. Sequences were submitted to GenBank under accession numbers KX383967-KX384018.
Sequence divergences were calculated using the K2P distance model (Kimura, 1980) following the barcoding approach suggested by Hebert et al. (2003; 2004). Phylogenetic analyses were made performing a Neighbour- Joining (NJ) tree with a distance matrix generated by using MEGA 6 (Tamura et al., 2013). Bootstrap values were obtained by making 1,000 replicates. Bayesian analyses were also performed using MrBayes (Ronquist and Huelsenbeck, 2003), by considering the best substitution models for each region using MrModelTest 3.7 (Nylander, 2004). Two independent runs of 2,000,000 generations were performed, with trees sampled every 100 generations, 25% of the trees were discarded as burning. Convergence was validated by the standard deviation of split frequencies (<0.01), by plotting the likelihood values over time, and by using the sump command in MrBayes. The percentage of trees recovering a particular clade was used as a measure of that clade’s posterior probability (Huelsenbeck and Ronquist, 2001).
RESULTS
Morphological analyses
Morphotype I
Description Seventy
adults with average weight 232.4±44.7 g, one specimen of 1 kg. Body is elongated, cylindrical, completed in rounded ends. The species presents varieties in body colorations (brown, reddish, yellow), size and in the ending of the body edges (Figure 2).

Figure 2. Isostichopus badionotus. a) Morphotype I. Typical morphotype, solid beige in the dorsal side, which is covered with dark chocolate round warts. b) Morphotype II. 1) Orange sea cucumber. The dorsal side is covered by orange warts that end in a dark color. 2) Dark sea cucumber surrounded by yellow round warts. 3) Ambulacral feet organized in three rows with podias that end in a dark color.
Specimens have brown coloration and numerous conical warts colouring brown. The dorsal side of the body presents a variable number of rectangular shape protuberances caused by depressions at the corners. This morphotype has an organization similar to four rows of low projections with small central spots that give it an appearance of breasts. In the two dorso-lateral margins there is only one row of warts and two more in the ventrolateral body. The ambulacral feet are of brown color with three lines of longitudinal podiums. Calcareous ring composed of projections in the form of pyramid and tower by way of a wheel, with a marked invagination at the base, with two small mountains or bumps on its outer edge. A respiratory tree and one or two poly vesicles are present. No presence of tubules of Cuvier. Gonads are placed in clusters, coloring whitecreamy, depending on the maturity gonad stage. Mature from July to November. Digestive tract is two times longer than body length, cylindrical beige, which occupies three times the length of their body. Ossicles: Towers, canes, in the shape of s and c, in badges perforated (Figure 3).
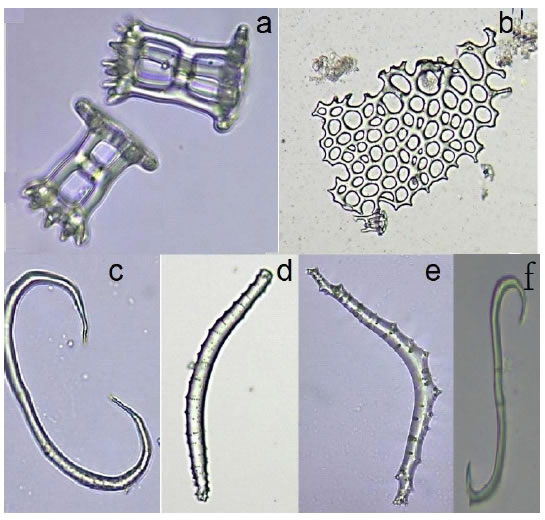
Figure 3. Isostichopus badionotus ossicles. a) Perforated tables, b) larger plates, c), d) and e) C-shaped rods, f) S-shaped rods.
Habitat (or ecology)
These specimens have been found in Colombia Caribbean Sea in Rodadero beach (11° 12′ 25.5″ N - 74° 13′ 54.8″ W), Airport beach (11° 09′ 20.2″ N - 74° 13′ 56.6″ W), and mainly in Taganga bay (11° 15′ 53.0″ N - 74° 11′ 32.0″ W). They inhabit sandy bottoms and have cryptic behavior. They feed on particulate material that includes algae, bacteria and marine sediment. Mature from July to November.
Morphotype II
Description
Adults with average weight 470.4±162.8 g, one specimen of 1 kg. Body is elongated, cylindrical, completed in rounded ends. These specimens present varieties in colorations (brown, reddish, yellow), size and in the ending of the edges (Figure 2). The body is light brown and on its surface has tiny pale spots with small, almost imperceptible dark papillae. They showed elongated body, cylindrical or fusiform, robust, with blunt edges. The ventral region is flat and the dorsal region is concave. The ventro-lateral margin is devoid of fleshy projections. The podiums are dark coffee arranged in three longitudinal rows, the middle one being thicker than those of the lateral ones. They have dark brown coloration with tiny conical pinnules, which colouring reddish-brown, whose endings are yellow in colour (Table 1). The mouth position is ventral and the anus, terminal. Eighteen to twenty peltates tentacles and anal opening without tooths. Presence of tube feet arranged in three rows, which become six rows when observed in the water. Their terminal colour is brown (Figure 2).
Table 1. Comparative characters among sea cucumber species Habitat (or ecology) These specimens have been found in the Colombia Caribbean Sea, mainly in Taganga bay (11° 15′ 53.0″ N - 74° 11′ 32.0″ W), inhabiting sandy bottoms. They feed on particulate material that includes algae, bacteria and marine sediment. Mature from July to November.
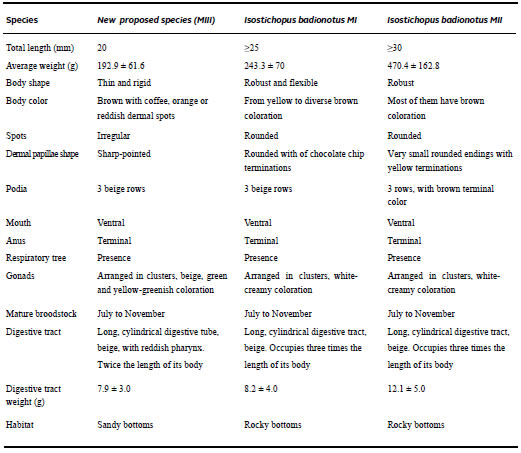
Calcareous ring composed of projections in the form of pyramid and tower by way of a wheel, with a marked invagination at the base, with two small mountains or bumps on its outer edge. A respiratory tree and one or two poly vesicles without tubules of Cuvier are present. Gonads are placed in clusters, coloring white-creamy, depending on the maturity gonad stage. Mature from July to November. Digestive tract long, three times longer than body length cylindrical beige, which occupies three times the length of their body. Ossicles: Towers, canes, in the shape of s and c, in badges perforated (Figure 3).
Morphotype III
Description
Adults with average weight around 192.9±61.6 g. Body is elongated, cylindrical, completed in blunt ends. The species presents varieties in colorations in the dorsal region, which are cream-yellow with irregular spots: coffee, orange or reddish. Its ventral region and the tube feet are beige (Figure 4) (Table 1).

Figure 4. New proposed species. a) Variations in irregular spots. Trapezoidal-shaped body. 1cm bar scale. b). Variations in body color. c) Close view of the peltates tentacles of Isostichopus isabellae n sp. d) Peltates tentacles contracted of the new proposed species. Lateral view. e) Ambulacral feet organized in three rows (arrows) with most of the podia ending in a cream color rather than in black (white line). Mouth (♦), pinules (*).
Mouth is ventral with 16-20, peltate tentacles, anus is terminal without teeth (Figure 4). Presence of ambulacral feet arranged in three rows. The dorsal region was slightly curved while the ventro-lateral margin of the wall was thick and provided with extensions called papillae. The calcareous ring has a pyramid form and tower with two small bumps on its top edge. Presence of respiratory tree. Presence of poly (1-2) vesicles without tubules of Cuvier. Gonads arranged in clusters, beige, green and yellow-greenish coloration, depending on the maturity gonad stage. Female gonads tubules reaching a diameter of 405.07±248.19 μm. Male gonads with thinner tubules which diameter was around of 272.18±56.37 μm. Mature oocyte diameter: 125.24±13.11 μm. Cuvierian organ absent. Long, cylindrical digestive tube of beige colour, with reddish pharynx, twice the length of its body (Figure 5).

Figure 5. Internal morphology of the new specie proposed. 1) Pigmented polian vesicle; 2) Respiratory tree; 3) Gonads; 4) Intestine; 5) Internal muscle; 6) Skin.
Ossicles: tables terminated in a crown, perforated by four central holes, c and s shaped rods, rounded and larger plates whit several holes (Figure 6).
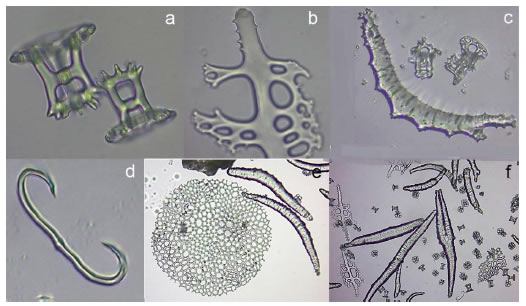
Figure 6. Ossicles of the dorsal body wall of the new specie proposed. a) Perforated tables with a single central hole, four smaller peripheral holes that end in crown as towers b) Rod with small holes, c) Rod of tentacles d) C-shaped rod, e) Perforate round plates of the dorsal body wall, f) Panoramic view of ossicles samples: very small tables, larger plates, rods some with holes.
Habitat (or ecology)
It inhabits rocky and sandy and feeds on particulate material that includes algae, bacteria and marine sediment. The species have been found in Rodadero beach, Airport beach, Taganga bay. Gonochoric species with bifurcated gonad attached to dorsal mesentery. Mature from July to November.
Remarks
Most of them are smaller in size than Morphotype I and II. They have a trapezoidal-shaped body, which is thin and rigid. They show dorsal irregular spots and different colours that varies from coffee, to red, orange and beige, depending on the individual. Conical pinnules. Ossicles are similar to that other Isostichopus. Seasonal spawning from July to November.
Sequence and phylogenetic analyses
Tables 2 and 3 show the divergent nucleotide sequences of the COI and 16S nucleotide alignments. For the COI gene three haplotypes corresponding to the three morphotypes where found (Table 2). The 16S gene is more variable, as expected, and we found more than one haplotype for morphotype III (Table 3). The results of our phylogenetic analyses are presented in Figure 7 and Figure 8. Both the NJ and the Bayesian analyses, for both genes (COI and 16S), recovered the same tree topology.
Table 2. Comparison of Santa Marta Isostichopus COI sequences from the three found morphotypes and other sequences available in Genbank. Numbers indicate divergent nucleotide positions along the alignment. Dots correspond to conserve nucleotides. Only sequences representative of each different haplotype are shown.
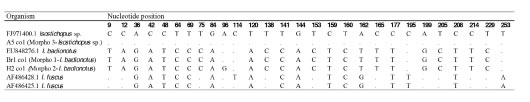
Table 3. Comparison of Santa Marta Isostichopus 16S sequences from the three found morphotypes and other sequences available in Genbank. Numbers indicate divergent nucleotide positions along the alignment. Dots correspond to conserve nucleotides. Only sequences representative of each different haplotype are shown.


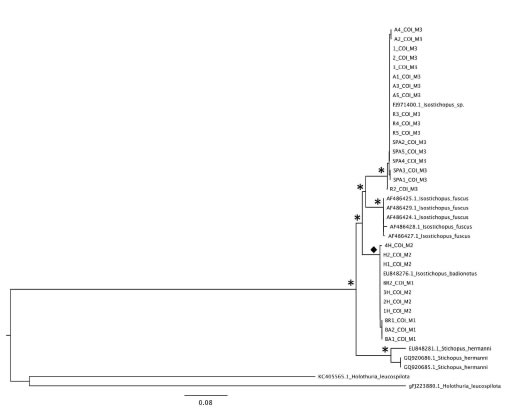
Figure 7. Tree topology obtained by Neighbor-joining and Bayesian analyses derived from COI (cytochrome c oxidase I) sequences showing two genetically distinct lineages of Isostichopus in Colombia. Sequences downloaded from GenBank are coded with each accession number. Asterisks represent posterior probabilities/booststraps that were > 0.90/90%, diamonds represent posterior probabilities/booststraps that were > 0.80/80%.

Figure 8. Tree topology obtained by Neighbor-joining and Bayesian analyses derived from 16S rRNA sequences showing two genetically distinct lineages of Isostichopus in Colombia. Sequences downloaded from GenBank are coded with each accession number. Asterisks represent posterior probabilities/booststraps that were > 0.90/90%, diamonds represent posterior probabilities/booststraps that were > 0.80/80%.
The phylogram obtained by Neighbor-joining and Bayesian analysis derived from COI sequences (Figure 7) showed that the three morphotypes of the so-called Isostichopus badionotus species form two well separated clades: one with all morphotype III (M3) individuals and the other including all morphotypes I (MI) and II (MII). The “morphotype III” clade also includes a sequence retrieved from GenBank identified as Isostichopus sp., the sister group of the “morphotype III” clade I. fuscus. The “morphotype I+II” clade also includes a sequence retrieved from GenBank identified as Isostichopus badionotus. This clade forms the sister group of the clade formed by morphotypes III and I. fuscus. The COI distance matrix indicated that the average distance percentage for Isostichopus was 0.20%, while the average between species distance value was 7.80%. Comparisons with S. herrmanni sequences indicated that the between genera distance average value was 16% (Table 2).
For the 16S distance matrix we obtained similar results, however, the sequence of I. macroparentheses available in Genbank is genetically very different from other Isostichopus sequences. In this case the average within species distance percentage for Isostichopus was of 0.5%, while the average between species distance value was of 13.78%. The distance values between I. macroparentheses and the other Isostichopus species were higher than the distance values obtained between the other Isostichopus species and Stichopus herrmanni (Table 3). One 16S sequence of I. badionotus (JN207495.1) clusters within the “Morphotype III” clade.
DISCUSSION
For years, there have been great controversy around the categorization of species worldwide and Colombian sea cucumber is no exception. Genetic studies are definitive for the understanding of the taxonomic identification of sea cucumbers. A species under a certain area can be easily confused or mistakenly classified if only its macroscopic characters are observed. There are pronounced differences between the two lineages studied in this work. One of the main features is that I. badionotus has strong chocolate spots with well pronounced edge, while Isostichopus sp. (morphotype III) presents different brownish spots distributed on the dorsal surface of the body. This morphotype was characterized by having a trapezoidalshaped body, which is in contrast to I. badionotus who has the ventral region flat and the dorsal region concave. Isostichopus sp. (morphotype III) was found always in rocky bottoms while I. badionotus was found in sandy bottoms as was state by Purcell et al., (2012). Additionally, the morphotype III digestive tract length, which occupies twice the length of its body in contrast to three times the length for I. badionotus. Our findings suggest morphotype III, could be a different species of Isostichopus.
Genetic evidence for taxonomic divergence
Since its appearance the DNA barcoding approach has been proposed as a strategy that could help on species characterization and discovery by allowing taxonomists to rapidly sort specimens and by highlighting divergent taxa that may represent new species (Hebert and Gregory, 2005). The increase in DNA-barcoding initiatives has led to the realization that, for a significant proportion of species, there is an actual correlation between genetic divergence and taxonomic status (Hebert et al., 2004). For the application of genetic barcodes we need to analyze the genetic divergence between and within species (interand intra-genetic distances), so that, for a given genetic marker (typically the COI gene), the lowest interspecific value does not overlap with the highest intraspecific value, maintaining a gap also called the “barcoding-gap” (Hebert et al., 2003). This threshold, depending on the organisms and the study, seems to lie around 2-4% (Hebert et al., 2003; Meyer and Paulay, 2005). Although the COI has proven to be a useful tool for the characterization of species in the Holothuroidea (Uthicke et al., 2010), the 16S rRNA gene has also been successfully tested for this purpose (Kamarudin, 2015; Wen and Zeng, 2014). In this study we evaluated the potential use of the COI and 16S rRNA genes for the characterization of Isostichopus species in Colombia.
In Uthicke´s et al. (2010) barcoding study with COI, conspecific sequence variation averages of 1.3% (numbers between 2.0 and 4.5%) and a congeneric average value of 16.9% were obtained, showing a clear “barcoding gap”. In our study, the average COI within-species distance value was of 0.20%, while the average between-species distance value was of 7.80%. We also obtained a clear “barcoding gap”. Comparisons with S. herrmanni sequences indicated that the average between-genera distance value was of 16%. Both for the COI and the 16S genes we found larger genetic distances values between Isostichopus sp. morphotype III and I. badionotus (morphotypes I and II) than between either of those with I. fuscus. Unfortunalty there is very little genetic information published in Genbank for the group. Although we tried to include the only 16S sequence available for I. macroparentheses, this sequence clearly does not belong to this species. When the sequence is compared to others in the dataset using the Blast tool, it matches another Istichopodid, Astichopus multifidus. Our tree reconstructions (both genes) showed the presence of two Isostichopus well differentiated groups. These barcoding results, corroborated by the morphological analyses, suggest that there might be two Isostichopus lineages or species in our Colombian samples. Furthermore, although we initially thought that the morphotype II could also be different to I. badionotus due to its external morphology, as proposed in Vergara and Rodriguez (2015), the genetic results revealed that it is part of the variety of morphotypes of the I. badionotus species. Nevertheless, its external characteristics, as well as the ossicles conformation directed the studies to analysis of genetic sequences that allowed us to confirm the presence of two lineages of Isostichopus in the Colombian Caribbean.
Isostichopus includes ten nominal Atlantic species: I. assimilis, I. maculatus, I. acanthomela, I. badionotus, I. diaboli, I. errans, I. haytiensis, I. macroparentheses, I. moebii and I. xanthomela (Heilprin, 1888). While in the present literature only two Atlantic species are recognized, it is quite possible that the morphotype III encountered here represents a described species currently relegated into the synonymy of I. badionotus. In order to confirm the taxonomical status of the Colombian Isostichopus, all these nominal species need to be evaluated on the basis of their descriptions and types. It is well known that studies that confirm the taxonomic classification of the species are an essential tool for the implementation of management plans and conservation programs for sea cucumbers. This study can be a tool to establish the plans that currently do not exist in the country.
ACKNOWLEDGMENTS
This work was supported by Vicerrectoria de Investigacion of the Universidad del Magdalena and Sistema General de Regalias Grant (Gobernacion del Magdalena-Convenio 090/2014-2015). Authors would like to thank Eng. Yahir Mendoza Vanegas and fisherman Jorge Polo who helped collecting holothurians and to the Grupo de Investigacion y Desarrollo Tecnologico en Acuicultura (GIDTA).
BIBLIOGRAPHY
1. Agudelo-Martinez V., Rodriguez-Forero A. (2017) Gametogenesis, spawning and larval development of Isostichopus sp. aff badionotus. SPC Beche-de-mer Information Bulletin 37: 65-74. [ Links ]
2. Agudelo V.Y., Rodriguez A. (2015) Advances on spontaneous captive breeding and culture conditions of Caribbean Sea cucumber Stichopus sp. SPC Bechede- Mer Information Bulletin 35: 50-57. [ Links ]
3. Arndt A., Marquez C., Lambert P., Smith M.J. (1996) Molecular phylogeny of eastern Pacific sea cucumbers (Echinodermata: Holothuroidea) based on mitochondrial DNA sequence. Mol. Phylogenet. Evol. 6: 425-437. [ Links ]
4. Borrero-Perez G., Benavides-Serrato M., Solano O., Navas G. (2003) Holothuroideos (Echinodermata: Holothuroidea) recolectados en el talud continental superior del Caribe colombiano. Boletin del Instituto Oceanografico de Venezuela. Universidad de Oriente 42: 65-85. [ Links ]
5. Borrero-Perez G.H., Benavides-Serrato M., Diaz-Sanchez C.M. (2012) Equinodermos del Caribe colombiano II: Echinoidea y Holothuroidea. Serie de Publicaciones Especiales de Invemar N° 30. [ Links ]
6. Bruckner, A. (2006). The proceedings of the technical workshop on the conservation of sea cucumbers in the families Holothuriidae and Stichopodidae. NOAA Technical Memorandum NMFS-OPR 44. NOAA, Silver Spring, MD. [ Links ]
7. Caycedo I.E. (1978) Holothurioidea (Echinodermata) de aguas someras en la costa norte de Colombia. Anales Inst. Invest. Mar. Punta Betin 10: 149-198. [ Links ]
8. Guzman H.M., Guevara C.A. (2002) Population structure, distribution and abundance of three commercial species of Sea Cucumber (Echinodermata) in Panama. Caribb. J. Sci. 38(3-4): 230-238. [ Links ]
9. Guzman, H., Guevara, C., Hernandez, I. (2003). Reproductive cycle of two commercial species of sea cucumber (Echinodermata: Holothuroidea) from Caribbean Panama. Marine Biology, 142(2), 271-279. [ Links ]
10. Hall T.A. (1999) BioEdit: a user-friendly biological sequences aligment editor and analysis program for Window 95/98/NT. Nucleic Acids Res. 41: 95-98. [ Links ]
11. Hebert P.D., Cywinska N.A., Ball S.L., de Waard J.R. (2003) Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B Biol. Sci. 270: 313-321. [ Links ]
12. Hebert P.D., Penton N., Burns J.M., Janzen D.H., Hallwachs W. (2004) Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes ulgerator. Proc. Natl. Acad. Sci. USA 101: 14812-14817. [ Links ]
13. Hebert P.D., Gregory T.R. (2005) The Promise of DNA Barcoding for Taxonomy. Syst. Biol. 54(5): 852-859. [ Links ]
14. Heilprin A. (1888) Contributions to the Natural History of the Bermuda Islands. P. Ac. Philad. pt. Iii, pp. 309-318. [ Links ]
15. Hendler G., Miller J.E., Pawson D.L., Kier P.M. (1995) Sea Stars, sea urchins and Allies: Echinoderms of Florida and the Caribbean. Smithsonian Institution Press. [ Links ]
16. Honey-Escandon M., Laguardia-Figueras A., Solis-Marin F.A. (2012) Molecular phylogeny of the subgenus Holothuria (Selenkothuria) Deichmann, 1958 (Holothurpoidea: Aspidochirotida). Zool. J. Linn. Soc. 165: 109-120. [ Links ]
17. Huelsenbeck J.P., Ronquist F. (2001) MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17: 754-755. [ Links ]
18. Kamarudin K.R. (2015) 16S rDNA Barcoding Technique for Molecular Identification of Processed Sea Cucumbers from Selected Malaysian Markets. Conference: IPCSM’15, At Convention Hall, E-Learning Building, Sultan Idris Education University (UPSI), Tanjong Malim, Perak Darul Ridzuan, MALAYSIA, pp. 22-26.
19. Kimura M. (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16: 111-120. [ Links ]
20. Laguarda-Figueras A. (2002) Equinodermos del Caribe de Mexico: Puerto Morelos, Quintana Roo. Universidad Nacional Autonoma de Mexico. Instituto de Ciencias del Mar y Limnologia. Informe final SNIBCONABIO proyectos N° S091. Mexico, D.F. [ Links ]
21. Lambert P. (1984) British Columbia marine faunistic survey report holothurians from the Northeast Pacific. Canadian Technical Reports of Fisheries and Aquatic Sciences 1234: 1-30. [ Links ]
22. Meyer C.P., Paulay G. (2005) DNA barcoding: error rates based on comprehensive sampling. Public Library of Science Biology 3 e422, pp. 10. [ Links ]
23. Miller, J. E., Pawson, David L. (1990). “Swimming sea cucumbers (Echinodermata: Holothuroidea): a survey, with analysis of swimming behavior in four bathyal species.” Smithsonian Contributions to the Marine Sciences. 1–18. https://doi.org/10.5479/si.01960768.35.1.
24. Nylander J. (2004) MrModeltest v2. Program distributed by the author. Evol. Biol. Uppsala University. [ Links ]
25. Pawson D.L. (2007) Phylum Echinodermata. In: Zhang Z.Q., Shear W.A. (Eds.) Linnaeus Tercentenary: Progress in Invertebrate Taxonomy. Zootaxa 1668: 1-766. [ Links ]
26. Purcell S.W., Samyn Y., Conand C. (2012) Commercially important sea cucumbers of the world. FAO. Species Catalogue for Fishery Purposes N° 6. [ Links ]
27. Rodriguez A., Agudelo V.Y., Vergara W.O. (2013) First insight into Colombian Caribbean sea cucumbers and sea cucumber fishery. SPC Beche-de-mer Information Bulletin 33: 9-13. [ Links ]
28. Ronquist F., Huelsenbeck J. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572-1574. [ Links ]
29. Smirnov A.V. (2012) System of the class Holothuroidea. Paleontol. J. 46(8): 793-832. [ Links ]
30. Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725-2729. [ Links ]
31. Tommasi L.R. (1969) Lista dos Holothuroidea recentes do Brasil. Contribuicoes Avulsas do Instituto Oceanografico, serie Oceanografia Biologica 15: 1-29. [ Links ]
32. Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., Higgins, D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 24, 4876-4882. [ Links ]
33. Toral-Granda V. (2008) Population status, fisheries and trade of sea cucumbers in Latin America and the Caribbean. Sea cucumbers. A global review of fisheries and trade. FAO Fisheries and Aquaculture Technical Paper 516: 213-229. [ Links ]
34. Toral-Granda V., Lovatelli A., Vasconcellos M. (2008) Sea cucumbers: A global review of fisheries and trade. FAO Fisheries and Aquaculture Technical Paper N° 516. Rome, FAO. [ Links ]
35. Uthicke S., Byrne M., Conand C. (2010) Genetic barcoding of commercial Beche-de-mer species (Echinodermata: Holothuroidea). Mol. Ecol. Res. 10: 634-646. [ Links ]
36. Vergara W., Rodriguez A. (2015) Histologia del tubo digestivo de tres especies de pepino de mar Isostichopus badionotus, Isostichopus sp. y Stichopus hermanni (Aspidochirotida: Stichopodidae). Rev. Biol. Trop. 63(4): 1021-1033. [ Links ]
37. Vergara W., Rodriguez A. (2016) Nutritional composition of sea cucumber Isostichopus sp. Nat. Resour. 7: 130- 137. [ Links ]
38. Wen J., Zeng L. (2014) Use of species-specific PCR for the identification of 10 sea cucumber species. Chin. J. Oceanol. Limnol. 32(6): 1257-1263. [ Links ]














